High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients
- PMID: 11283081
- PMCID: PMC87964
- DOI: 10.1128/JCM.39.4.1522-1529.2001
High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients
Abstract
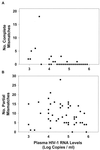
We assessed the reproducibility of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and protease sequencing using cryopreserved plasma aliquots obtained from 46 heavily treated HIV-1-infected individuals in two laboratories using dideoxynucleotide sequencing. The rates of complete sequence concordance between the two laboratories were 99.1% for the protease sequence and 99.0% for the RT sequence. Approximately 90% of the discordances were partial, defined as one laboratory detecting a mixture and the second laboratory detecting only one of the mixture's components. Only 0.1% of the nucleotides were completely discordant between the two laboratories, and these were significantly more likely to occur in plasma samples with lower plasma HIV-1 RNA levels. Nucleotide mixtures were detected at approximately 1% of the nucleotide positions, and in every case in which one laboratory detected a mixture, the second laboratory either detected the same mixture or detected one of the mixture's components. The high rate of concordance in detecting mixtures and the fact that most discordances between the two laboratories were partial suggest that most discordances were caused by variation in sampling of the HIV-1 quasispecies by PCR rather than by technical errors in the sequencing process itself.
Figures





References
-
- Baxter J D, Mayers D L, Wentworth D N, Neaton J D, Hoover M L, Winters M A, Mannheimer S B, Thompson M A, Abrams D I, Brizz B J, Ioannidis J P, Merigan T C. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–F93. - PubMed
-
- Carpenter C C, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society-USA Panel. JAMA. 2000;283:381–390. - PubMed
-
- D'Aquila R T. Limits of resistance testing. Antivir Ther. 2000;5:71–76. - PubMed
-
- DeGruttola V, Dix L, D'Aquila R, Holder D, Phillips A, Ait-Khaled M, Baxter J, Clevenbergh P, Hammer S, Harrigan R, Katzenstein D, Lanier R, Miller M, Para M, Yerly S, Zolopa A, Murray J, Patick A, Miller V, Castillo S, Pedneault L, Mellors J. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther. 2000;5:41–48. - PubMed
-
- Demeter L M, D'Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard T M, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working Group. AIDS Clinical Trials Group. J Virol Methods. 1998;75:93–104. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

