Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific
- PMID: 8917550
- PMCID: PMC24052
- DOI: 10.1073/pnas.93.23.13096
Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific
Abstract
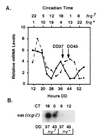
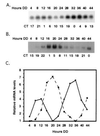
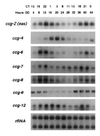
An endogenous circadian biological clock controls the temporal aspects of life in most organisms, including rhythmic control of genes involved in clock output pathways. In the fungus Neurospora crassa, one pathway known to be under control of the clock is asexual spore (conidia) development. To understand more fully the processes that are regulated by the N. crassa circadian clock, systematic screens were carried out for genes that oscillate at the transcriptional level. Time-of-day-specific cDNA libraries were generated and used in differential screens to identify six new clock-controlled genes (ccgs). Transcripts specific for each of the ccgs preferentially accumulate during the late night to early morning, although they vary with respect to steady-state mRNA levels and amplitude of the rhythm. Sequencing of the ends of the new ccg cDNAs revealed that ccg-12 is identical to N. crassa cmt encoding copper metallothionein, providing the suggestion that not all clock-regulated genes in N. crassa are specifically involved in the development of conidia. This was supported by finding that half of the new ccgs, including cmt(ccg-12), are not transcriptionally induced by developmental or light signals. These data suggest a major role for the clock in the regulation of biological processes distinct from development.
Figures


References
-
- Edmunds L N. Cellular and Molecular Bases of Biological Clocks. New York: Springer; 1988.
-
- Loros J J. Semin Neurosci. 1995;7:3–13.
-
- Lakin-Thomas P L, Coté G G, Brody S. Crit Rev Microbiol. 1990;17:365–416. - PubMed
-
- Loros J J, Denome S A, Dunlap J C. Science. 1989;243:385–388. - PubMed
-
- Liu Y, Tsinoremas N F, Johnson C H, Levedeva N V, Golden S S, Ishiura M, Kondo T. Genes Dev. 1995;9:1469–1478. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

