An imprinted, mammalian bicistronic transcript encodes two independent proteins
- PMID: 10318933
- PMCID: PMC21909
- DOI: 10.1073/pnas.96.10.5616
An imprinted, mammalian bicistronic transcript encodes two independent proteins
Abstract
Polycistronic transcripts are common in prokaryotes but rare in eukaryotes. Phylogenetic analysis of the SNRPN (SmN) mRNA in five eutherian mammals reveals a second highly conserved coding sequence, termed SNURF (SNRPN upstream reading frame). The vast majority of nucleotide substitutions in SNURF occur in the wobble codon position, providing strong evolutionary evidence for selection for protein-coding function. Because SNURF-SNRPN maps to human chromosome 15q11-q13 and is paternally expressed, each cistron is a candidate for a role in the imprinted Prader-Willi syndrome (PWS) and PWS mouse models. SNURF encodes a highly basic 71-aa protein that is nuclear-localized (as is SmN). Because SNURF is the only protein-coding sequence within the imprinting regulatory region in 15q11-q13, it may have provided the original selection for imprinting in this domain. Whereas some human tissues express a minor SNURF-only transcript, mouse tissues express only the bicistronic Snurf-Snrpn transcript. We show that both SNURF and SNRPN are translated in normal, but not PWS, human, and mouse tissues and cell lines. These findings identify SNURF as a protein that is produced along with SmN from a bicistronic transcript; polycistronic mRNAs therefore are encoded in mammalian genomes where they may form functional operons.
Figures


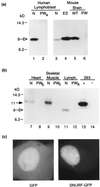

References
-
- Bartolomei M S, Tilghman S M. Annu Rev Genet. 1997;31:493–525. - PubMed
-
- Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. - PubMed
-
- Schmauss C, Brines M L, Lerner M R. J Biol Chem. 1992;267:8521–8529. - PubMed
-
- Sutcliffe J S, Nakao M, Christian S, Örstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Nat Genet. 1994;8:52–58. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

