Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya
- PMID: 10196337
- PMCID: PMC104220
- DOI: 10.1128/JVI.73.5.4393-4403.1999
Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya
Abstract
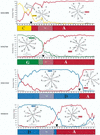
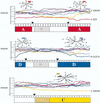
In sub-Saharan Africa, where the effects of human immunodeficiency virus type 1 (HIV-1) have been most devastating, there are multiple subtypes of this virus. The distribution of different subtypes within African populations is generally not linked to particular risk behaviors. Thus, Africa is an ideal setting in which to examine the diversity and mixing of viruses from different subtypes on a population basis. In this setting, it is also possible to address whether infection with a particular subtype is associated with differences in disease stage. To address these questions, we analyzed the HIV-1 subtype, plasma viral loads, and CD4 lymphocyte levels in 320 women from Nairobi, Kenya. Subtype was determined by a combination of heteroduplex mobility assays and sequence analyses of envelope genes, using geographically diverse subtype reference sequences as well as envelope sequences of known subtype from Kenya. The distribution of subtypes in this population was as follows: subtype A, 225 (70.3%); subtype D, 65 (20.5%); subtype C, 22 (6.9%); and subtype G, 1 (0.3%). Intersubtype recombinant envelope genes were detected in 2.2% of the sequences analyzed. Given that the sequences analyzed represented only a small fraction of the proviral genome, this suggests that intersubtype recombinant viral genomes may be very common in Kenya and in other parts of Africa where there are multiple subtypes. The plasma viral RNA levels were highest in women infected with subtype C virus, and women infected with subtype C virus had significantly lower CD4 lymphocyte levels than women infected with the other subtypes. Together, these data suggest that women in Kenya who are infected with subtype C viruses are at more advanced stages of immunosuppression than women infected with subtype A or D. There are at least two models to explain the data from this cross-sectional study; one is that infection with subtype C is associated with a more rapid disease progression, and the second is that subtype C represents an older epidemic in Kenya. Discriminating between these possibilities in a longitudinal study will be important for increasing our understanding of the role of specific subtypes in the transmission and pathogenesis of HIV-1.
Figures



References
-
- Bodrug S, Domingo R, Holloway J, Sanders M, Nunomura K, Sloan C, Billyard B. Gen-Probe single-tube quantitative HIV assay. J Clin Microbiol Infect. 1997;3:1050.
-
- Burke D S, McCutchan F E. Global distribution of human immunodeficiency virus-1 clades. In: Vincent S H, DeVita T Jr, Rosenberg Steven A, editors. AIDS: biology, diagnosis, treatment and prevention. Vol. 4. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 119–126.
-
- Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–354. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

