Rapid and systemic accumulation of chloroplast mRNA-binding protein transcripts after flame stimulus in tomato
- PMID: 10517843
- PMCID: PMC59414
- DOI: 10.1104/pp.121.2.517
Rapid and systemic accumulation of chloroplast mRNA-binding protein transcripts after flame stimulus in tomato
Abstract
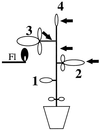
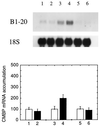
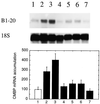
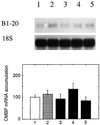
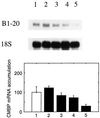
It has been shown that tomato (Lycopersicon esculentum) plants respond to flame wounding and electrical stimulation by a rapid (15 min) and systemic up-regulation of proteinase inhibitor (pin) genes. To find other genes having a similar expression pattern, we used subtractive cDNA screening between flamed and control plants to select clones up-regulated by flame wounding. We report the characterization of one of them, a chloroplast mRNA-binding protein encoded by a single gene and expressed preferentially in the leaves. Systemic gene expression in response to flaming in the youngest terminal leaf exhibited three distinct phases: a rapid and transient increase (5-15 min) in transcript accumulation, a decline to basal levels (15-45 min), and then a second, more prolonged increase (60-90 min). In contrast, after a mechanical wound the rapid, transient increase (5 min) was followed by a rapid decline to basal levels but no later, prolonged accumulation. In the petiole, the initial flame-wound-evoked transient increase (15 min) was followed by a continuous decline for 3 h. The nature of the wound signal(s) causing such rapid changes in transcript abundance is discussed in relation to electrical signaling, which has recently been implicated in plant responses to wounding.
Figures


References
-
- Bonaldo M de F, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. - PubMed
-
- Braam J, Davis R W. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin related genes in Arabidopsis. Cell. 1990;60:357–364. - PubMed
-
- Christensen AH, Sharrok RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

