cpmA, a gene involved in an output pathway of the cyanobacterial circadian system
- PMID: 10348865
- PMCID: PMC93820
- DOI: 10.1128/JB.181.11.3516-3524.1999
cpmA, a gene involved in an output pathway of the cyanobacterial circadian system
Abstract
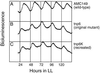
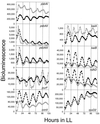
We generated random mutations in Synechococcus sp. strain PCC 7942 to look for genes of output pathways in the cyanobacterial circadian system. A derivative of transposon Tn5 was introduced into the chromosomes of reporter strains in which cyanobacterial promoters drive the Vibrio harveyi luxAB genes and produce an oscillation of bioluminescence as a function of circadian gene expression. Among low-amplitude mutants, one mutant, tnp6, had an insertion in a 780-bp open reading frame. The tnp6 mutation produced an altered circadian phasing phenotype in the expression rhythms of psbAI::luxAB, psbAII::luxAB, and kaiA::luxAB but had no or little effect on those of psbAIII::luxAB, purF::luxAB, kaiB::luxAB, rpoD2::luxAB, ndhD::luxAB, and conII::luxAB. This suggests that the interrupted gene in tnp6, named cpmA (circadian phase modifier), is part of a circadian output pathway that regulates the expression rhythms of psbAI, psbAII, and kaiA.
Figures



References
-
- Andersson, C. A., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. Min, and S. S. Golden. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol., in press. - PubMed
-
- Aschoff J. Temporal orientation: circadian clocks in animals and humans. Anim Behav. 1989;37:881–896.
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987.
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

