Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean
- PMID: 10388721
- PMCID: PMC91474
- DOI: 10.1128/AEM.65.7.3192-3204.1999
Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean
Abstract
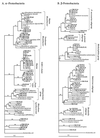
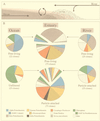
The Columbia River estuary is a dynamic system in which estuarine turbidity maxima trap and extend the residence time of particles and particle-attached bacteria over those of the water and free-living bacteria. Particle-attached bacteria dominate bacterial activity in the estuary and are an important part of the estuarine food web. PCR-amplified 16S rRNA genes from particle-attached and free-living bacteria in the Columbia River, its estuary, and the adjacent coastal ocean were cloned, and 239 partial sequences were determined. A wide diversity was observed at the species level within at least six different bacterial phyla, including most subphyla of the class Proteobacteria. In the estuary, most particle-attached bacterial clones (75%) were related to members of the genus Cytophaga or of the alpha, gamma, or delta subclass of the class Proteobacteria. These same clones, however, were rare in or absent from either the particle-attached or the free-living bacterial communities of the river and the coastal ocean. In contrast, about half (48%) of the free-living estuarine bacterial clones were similar to clones from the river or the coastal ocean. These free-living bacteria were related to groups of cosmopolitan freshwater bacteria (beta-proteobacteria, gram-positive bacteria, and Verrucomicrobium spp.) and groups of marine organisms (gram-positive bacteria and alpha-proteobacteria [SAR11 and Rhodobacter spp.]). These results suggest that rapidly growing particle-attached bacteria develop into a uniquely adapted estuarine community and that free-living estuarine bacteria are similar to members of the river and the coastal ocean microbial communities. The high degree of diversity in the estuary is the result of the mixing of bacterial communities from the river, estuary, and coastal ocean.
Figures





References
-
- Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277.
-
- Baross J A, Crump B, Simenstad C A. Elevated ‘microbial loop’ activities in the Columbia River estuarine turbidity maximum. In: Dyer K R, Orth B J, editors. Changes in fluxes in estuaries: implications from science to management (ECSA22/ERF symposium, Plymouth, September 1992). Fredensborg, Denmark: Olsen & Olsen; 1994. pp. 495–464.
-
- Berner E K, Berner R A. Global environment: water, air, and geochemical cycles. Englewood Cliffs, N.J: Prentice Hall; 1996. Marginal marine environments: estuaries; pp. 284–311.
-
- Crump B C, Baross J A. Particle-attached bacteria and heterotrophic plankton in the Columbia River estuary. Mar Ecol Prog Ser. 1996;138:265–273.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

