The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons
- PMID: 10762254
- PMCID: PMC111316
- DOI: 10.1128/JB.182.9.2520-2529.2000
The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons
Abstract
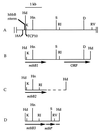
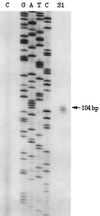
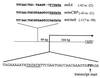
Three different methyltransferases initiate methanogenesis from trimethylamine (TMA), dimethylamine (DMA) or monomethylamine (MMA) by methylating different cognate corrinoid proteins that are subsequently used to methylate coenzyme M (CoM). Here, genes encoding the DMA and TMA methyltransferases are characterized for the first time. A single copy of mttB, the TMA methyltransferase gene, was cotranscribed with a copy of the DMA methyltransferase gene, mtbB1. However, two other nearly identical copies of mtbB1, designated mtbB2 and mtbB3, were also found in the genome. A 6.8-kb transcript was detected with probes to mttB and mtbB1, as well as to mtbC and mttC, encoding the cognate corrinoid proteins for DMA:CoM and TMA:CoM methyl transfer, respectively, and with probes to mttP, encoding a putative membrane protein which might function as a methylamine permease. These results indicate that these genes, found on the chromosome in the order mtbC, mttB, mttC, mttP, and mtbB1, form a single transcriptional unit. A transcriptional start site was detected 303 or 304 bp upstream of the translational start of mtbC. The MMA, DMA, and TMA methyltransferases are not homologs; however, like the MMA methyltransferase gene, the genes encoding the DMA and TMA methyltransferases each contain a single in-frame amber codon. Each of the three DMA methyltransferase gene copies from Methanosarcina barkeri contained an amber codon at the same position, followed by a downstream UAA or UGA codon. The C-terminal residues of DMA methyltransferase purified from TMA-grown cells matched the residues predicted for the gene products of mtbB1, mtbB2, or mtbB3 if termination occurred at the UAA or UGA codon rather than the in-frame amber codon. The mttB gene from Methanosarcina thermophila contained a UAG codon at the same position as the M. barkeri mttB gene. The UAG codon is also present in mttB transcripts. Thus, the genes encoding the three types of methyltransferases that initiate methanogenesis from methylamine contain in-frame amber codons that are suppressed during expression of the characterized methyltransferases.
Figures




References
-
- Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. - PubMed
-
- Boone D R, Whitman W B, Rouviére P. Diversity and taxonomy of methanogens. In: Ferry J G, editor. Methanogenesis. Ecology, physiology, biochemistry, and genetics. New York, N.Y: Chapman & Hall; 1993. pp. 35–80.
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
-
- Burke S A, Krzycki J A. Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J Biol Chem. 1997;272:16570–16577. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

