Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia
- PMID: 10449476
- PMCID: PMC85414
- DOI: 10.1128/JCM.37.9.2920-2926.1999
Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia
Abstract
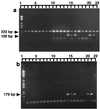
Cryptococcus neoformans var. gattii lives in association with certain species of eucalyptus trees and is a causative agent of cryptococcosis. It exists as two mating types, MATalpha and MATa, which is determined by a single-locus, two-allele system. In the closely related C. neoformans var. neoformans, the alpha mating type has been found to outnumber its a counterpart by at least 30:1, but there have been very limited data on the proportions of each mating type in C. neoformans var. gattii. In the present study, specific PCR primers were designed to amplify two separate alpha-mating-type genes from C. neoformans var. gattii strains. These were used to survey for the presence of the two mating types in clinical and environmental collections of C. neoformans var. gattii strains from Australia. Sixty-eight of 69 clinical isolates produced both alpha mating type-specific bands and were assumed to be of the alpha mating type. The majority of environmental isolates were also of the alpha mating type, but the a mating type was located in two separate areas. In one area, the a mating type outnumbered the alpha mating type by 27:2, but in the second area, the ratio of the two mating types was close to the 50:50 ratio expected for sexual recombination.
Figures


References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

