The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts
- PMID: 14681369
- PMCID: PMC308836
- DOI: 10.1093/nar/gkh102
The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts
Abstract
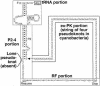
tmRNA combines tRNA- and mRNA-like properties and ameliorates problems arising from stalled ribosomes. Research on the mechanism, structure and biology of tmRNA is served by the tmRNA website (http://www.indiana.edu/~ tmrna), a collection of sequences, alignments, secondary structures and other information. Because many of these sequences are not in GenBank, a BLAST server has been added; another new feature is an abbreviated alignment for the tRNA-like domain only. Many tmRNA sequences from plastids have been added, five found in public sequence data and another 10 generated by direct sequencing; detection in early-branching members of the green plastid lineage brings coverage to all three primary plastid lineages. The new sequences include the shortest known tmRNA sequence. While bacterial tmRNAs usually have a lone pseudoknot upstream of the mRNA segment and a string of three or four pseudoknots downstream, plastid tmRNAs collectively show loss of pseudoknots at both postions. The pseudoknot-string region is also too short to contain the usual pseudoknot number in another new entry, the tmRNA sequence from a bacterial endosymbiont of insect cells, Tremblaya princeps. Pseudoknots may optimize tmRNA function in free-living bacteria, yet become dispensible when the endosymbiotic lifestyle relaxes selective pressure for fast growth.
Figures
References
-
- Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials