Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes
- PMID: 10570164
- PMCID: PMC24156
- DOI: 10.1073/pnas.96.24.13863
Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes
Abstract
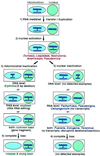
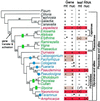
The respiratory gene cox2, normally present in the mitochondrion, was previously shown to have been functionally transferred to the nucleus during flowering plant evolution, possibly during the diversification of legumes. To search for novel intermediate stages in the process of intracellular gene transfer and to assess the evolutionary timing and frequency of cox2 transfer, activation, and inactivation, we examined nuclear and mitochondrial (mt) cox2 presence and expression in over 25 legume genera and mt cox2 presence in 392 genera. Transfer and activation of cox2 appear to have occurred during recent legume evolution, more recently than previously inferred. Many intermediate stages of the gene transfer process are represented by cox2 genes in the studied legumes. Nine legumes contain intact copies of both nuclear and mt cox2, although transcripts could not be detected for some of these genes. Both cox2 genes are transcribed in seven legumes that are phylogenetically interspersed with species displaying only nuclear or mt cox2 expression. Inactivation of cox2 in each genome has taken place multiple times and in a variety of ways, including loss of detectable transcripts or transcript editing and partial to complete gene loss. Phylogenetic evidence shows about the same number (3-5) of separate inactivations of nuclear and mt cox2, suggesting that there is no selective advantage for a mt vs. nuclear location of cox2 in plants. The current distribution of cox2 presence and expression between the nucleus and mitochondrion in the studied legumes is probably the result of chance mutations silencing either cox2 gene.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

