Genetic diversity among Lassa virus strains
- PMID: 10888638
- PMCID: PMC112216
- DOI: 10.1128/jvi.74.15.6992-7004.2000
Genetic diversity among Lassa virus strains
Abstract
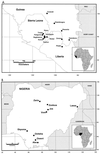
The arenavirus Lassa virus causes Lassa fever, a viral hemorrhagic fever that is endemic in the countries of Nigeria, Sierra Leone, Liberia, and Guinea and perhaps elsewhere in West Africa. To determine the degree of genetic diversity among Lassa virus strains, partial nucleoprotein (NP) gene sequences were obtained from 54 strains and analyzed. Phylogenetic analyses showed that Lassa viruses comprise four lineages, three of which are found in Nigeria and the fourth in Guinea, Liberia, and Sierra Leone. Overall strain variation in the partial NP gene sequence was found to be as high as 27% at the nucleotide level and 15% at the amino acid level. Genetic distance among Lassa strains was found to correlate with geographic distance rather than time, and no evidence of a "molecular clock" was found. A method for amplifying and cloning full-length arenavirus S RNAs was developed and used to obtain the complete NP and glycoprotein gene (GP1 and GP2) sequences for two representative Nigerian strains of Lassa virus. Comparison of full-length gene sequences for four Lassa virus strains representing the four lineages showed that the NP gene (up to 23.8% nucleotide difference and 12.0% amino acid difference) is more variable than the glycoprotein genes. Although the evolutionary order of descent within Lassa virus strains was not completely resolved, the phylogenetic analyses of full-length NP, GP1, and GP2 gene sequences suggested that Nigerian strains of Lassa virus were ancestral to strains from Guinea, Liberia, and Sierra Leone. Compared to the New World arenaviruses, Lassa and the other Old World arenaviruses have either undergone a shorter period of diverisification or are evolving at a slower rate. This study represents the first large-scale examination of Lassa virus genetic variation.
Figures


References
-
- Albarino C G, Ghiringhelli P D, Posik D M, Lozano M E, Ambrosio A M, Sanchez A, Romanowski V. Molecular characterization of attenuated Junin virus strains. J Gen Virol. 1997;78:1605–1610. - PubMed
-
- Albarino C G, Posik D M, Ghiringhelli P D, Lozano M E, Romanowski V. Arenavirus phylogeny: a new insight. Virus Genes. 1998;16:39–46. - PubMed
-
- Anonymous. Lassa fever. Wkly Epidemiol Rec. 1997;20:145–146.
-
- Auperin D D, Galinski M, Bishop D H. The sequences of the N protein gene and intergenic region of the S RNA of Pichinde arenavirus. Virology. 1984;134:208–219. - PubMed
-
- Auperin D D, McCormick J B. Nucleotide sequence of the Lassa virus (Josiah strain) S genome RNA and amino acid sequence comparison of the N and GPC proteins to other arenaviruses. Virology. 1989;168:421–425. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

