Evolutionarily conserved human targets of adenosine to inosine RNA editing
- PMID: 15731336
- PMCID: PMC549564
- DOI: 10.1093/nar/gki239
Evolutionarily conserved human targets of adenosine to inosine RNA editing
Abstract
A-to-I RNA editing by ADARs is a post-transcriptional mechanism for expanding the proteomic repertoire. Genetic recoding by editing was so far observed for only a few mammalian RNAs that are predominantly expressed in nervous tissues. However, as these editing targets fail to explain the broad and severe phenotypes of ADAR1 knockout mice, additional targets for editing by ADARs were always expected. Using comparative genomics and expressed sequence analysis, we identified and experimentally verified four additional candidate human substrates for ADAR-mediated editing: FLNA, BLCAP, CYFIP2 and IGFBP7. Additionally, editing of three of these substrates was verified in the mouse while two of them were validated in chicken. Interestingly, none of these substrates encodes a receptor protein but two of them are strongly expressed in the CNS and seem important for proper nervous system function. The editing pattern observed suggests that some of the affected proteins might have altered physiological properties leaving the possibility that they can be related to the phenotypes of ADAR1 knockout mice.
Figures

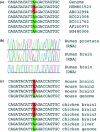

References
-
- Polson A.G., Crain P.F., Pomerantz S.C., McCloskey J.A., Bass B.L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991;30:11507–11514. - PubMed
-
- Wang Q., Khillan J., Gadue P., Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. - PubMed
-
- Palladino M.J., Keegan L.P., O'Connell M.A., Reenan R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. - PubMed
-
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

