Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes
- PMID: 11114919
- PMCID: PMC94868
- DOI: 10.1128/JB.183.1.214-220.2001
Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes
Abstract
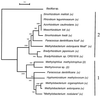
Rhizobia described so far belong to three distinct phylogenetic branches within the alpha-2 subclass of Proteobacteria. Here we report the discovery of a fourth rhizobial branch involving bacteria of the Methylobacterium genus. Rhizobia isolated from Crotalaria legumes were assigned to a new species, "Methylobacterium nodulans," within the Methylobacterium genus on the basis of 16S ribosomal DNA analyses. We demonstrated that these rhizobia facultatively grow on methanol, which is a characteristic of Methylobacterium spp. but a unique feature among rhizobia. Genes encoding two key enzymes of methylotrophy and nodulation, the mxaF gene, encoding the alpha subunit of the methanol dehydrogenase, and the nodA gene, encoding an acyltransferase involved in Nod factor biosynthesis, were sequenced for the type strain, ORS2060. Plant tests and nodA amplification assays showed that "M. nodulans" is the only nodulating Methylobacterium sp. identified so far. Phylogenetic sequence analysis showed that "M. nodulans" NodA is closely related to Bradyrhizobium NodA, suggesting that this gene was acquired by horizontal gene transfer.
Figures




References
-
- Allen O N, Allen E K. The Leguminosae, a source book of characteristics, uses, and nodulation. Madison: University of Wisconsin Press; 1981.
-
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microbiol Physiol. 1986;27:113–210. - PubMed
-
- Becker M, Johnson D E. The role of legume fallows in intensified upland rice-based systems of West Africa. Nutr Cycling Agroecosyst. 1999;1:71–81.
-
- Belgian Co-ordinated Collection of Micro-organisms/Laboratorium voor Microbiologie. Bacteria. Catalogue. Ghent, Belgium: Universiteit Gent; 1998.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

