Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences
- PMID: 11571190
- PMCID: PMC93237
- DOI: 10.1128/AEM.67.10.4828-4833.2001
Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences
Abstract
Streptococcus suis is an important pathogen of swine which occasionally infects humans as well. There are 35 serotypes known for this organism, and it would be desirable to develop rapid methods methods to identify and differentiate the strains of this species. To that effect, partial chaperonin 60 gene sequences were determined for the 35 serotype reference strains of S. suis. Analysis of a pairwise distance matrix showed that the distances ranged from 0 to 0.275 when values were calculated by the maximum-likelihood method. For five of the strains the distances from serotype 1 were greater than 0.1, and for two of these strains the distances were were more than 0.25, suggesting that they belong to a different species. Most of the nucleotide differences were silent; alignment of protein sequences showed that there were only 11 distinct sequences for the 35 strains under study. The chaperonin 60 gene phylogenetic tree was similar to the previously published tree based on 16S rRNA sequences, and it was also observed that strains with identical chaperonin 60 gene sequences tended to have identical 16S rRNA sequences. The chaperonin 60 gene sequences provided a higher level of discrimination between serotypes than the 16S RNA sequences provided and could form the basis for a diagnostic protocol.
Figures
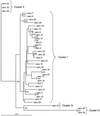
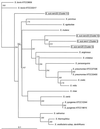


References
-
- Beaudoin M, Harel J, Higgins R, Gottschalk M, Frenette M, MacInnes J I. Molecular analysis of isolates of Streptococcus suis capsular type 2 by restriction-endonuclease-digested DNA separated on SDS-PAGE and by hybridization with an rDNA probe. J Gen Microbiol. 1992;138:2639–2645. - PubMed
-
- Boye M, Feenstra A A, Tegtmeier C, Andresen L O, Rasmussen S R, Bille-Hansen V. Detection of Streptococcus suis by in situ hybridization, indirect immunofluorescence, and peroxidase-antiperoxidase assays in formalin-fixed, paraffin-embedded tissue sections from pigs. J Vet Diagn Invest. 2000;12:224–232. - PubMed
-
- Chanter N, Jones P W, Alexander T J. Meningitis in pigs caused by Streptococcus suis — a speculative review. Vet Microbiol. 1993;36:39–55. - PubMed
-
- Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese L A, Brousseau R. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48:581–589. - PubMed
-
- Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. 5, suppl. 3. Washington, D.C.: National Biomedical Research Foundation; 1978. pp. 345–352.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

