Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability
- PMID: 10877758
- PMCID: PMC92063
- DOI: 10.1128/AEM.66.7.2703-2710.2000
Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability
Abstract
Reduced bioavailability of nonpolar contaminants due to sorption to natural organic matter is an important factor controlling biodegradation of pollutants in the environment. We established enrichment cultures in which solid organic phases were used to reduce phenanthrene bioavailability to different degrees (R. J. Grosser, M. Friedrich, D. M. Ward, and W. P. Inskeep, Appl. Environ. Microbiol. 66:2695-2702, 2000). Bacteria enriched and isolated from contaminated soils under these conditions were analyzed by denaturing gradient gel electrophoresis (DGGE) and sequencing of PCR-amplified 16S ribosomal DNA segments. Compared to DGGE patterns obtained with enrichment cultures containing sand or no sorptive solid phase, different DGGE patterns were obtained with enrichment cultures containing phenanthrene sorbed to beads of Amberlite IRC-50 (AMB), a weak cation-exchange resin, and especially Biobead SM7 (SM7), a polyacrylic resin that sorbed phenanthrene more strongly. SM7 enrichments selected for mycobacterial phenanthrene mineralizers, whereas AMB enrichments selected for a Burkholderia sp. that degrades phenanthrene. Identical mycobacterial and Burkholderia 16S rRNA sequence segments were found in SM7 and AMB enrichment cultures inoculated with contaminated soil from two geographically distant sites. Other closely related Burkholderia sp. populations, some of which utilized phenanthrene, were detected in sand and control enrichment cultures. Our results are consistent with the hypothesis that different phenanthrene-utilizing bacteria inhabiting the same soils may be adapted to different phenanthrene bioavailabilities.
Figures

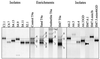

References
-
- Achouak W, Christen R, Barakat M, Martel M H, Heulin T. Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int J Syst Bacteriol. 1999;49:787–794. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

