Phylogenetic analysis of Lacazia loboi places this previously uncharacterized pathogen within the dimorphic Onygenales
- PMID: 11136789
- PMCID: PMC87720
- DOI: 10.1128/JCM.39.1.309-314.2001
Phylogenetic analysis of Lacazia loboi places this previously uncharacterized pathogen within the dimorphic Onygenales
Abstract
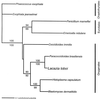
Lacazia loboi is the last of the classical fungal pathogens to remain a taxonomic enigma, primarily because it has resisted cultivation and only causes cutaneous and subcutaneous infections in humans and dolphins in the New World tropics. To place it in the evolutionary tree of life, as has been done for the other enigmatic human pathogens Pneumocystis carinii and Rhinosporidium seeberi, we amplified its 18S small-subunit ribosomal DNA (SSU rDNA) and 600 bp of its chitin synthase-2 gene. Our phylogenetic analysis indicated that L. loboi is the sister taxon of the human dimorphic fungal pathogen Paracoccidioides brasiliensis and that both species belong with the other dimorphic fungal pathogens in the order Onygenales. The low nucleotide variation among three P. brasiliensis 18S SSU rDNA sequences contrasts with the surprising amount of nucleotide differences between the two sequences of L. loboi used in this study, suggesting that the nucleic acid epidemiology of this hydrophilic pathogen will be rewarding.
Figures


References
-
- Ajello L. Ecology and epidemiology of hydrophilic infectious fungi and parafungi of medical mycological importance: a new category of pathogens. In: Ajello L, Hay R J, editors. Topley & Wilson's microbiology and microbial infections, medical mycology. 9th ed. Vol. 4. London, England: Arnold; 1998. pp. 67–73.
-
- Borelli D. Lobomicose: nomen de su agente (revision crìtica) Med Cut. 1968;3:151–156.
-
- Bowman B, Taylor J W, White T J. Molecular evolution of the fungi: human pathogens. Mol Biol Evol. 1992;9:893–904. - PubMed
-
- Bowman B H, Taylor J W. Molecular phylogeny of pathogenic and non-pathogenic Onygenales. In: Reynolds D R, Taylor J W, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, United Kingdom: CAB International; 1993. pp. 169–178.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

