Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm
- PMID: 12426392
- PMCID: PMC137205
- DOI: 10.1093/emboj/cdf613
Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm
Abstract
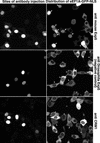
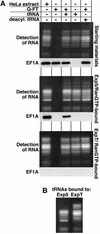
Importin beta-type transport receptors mediate the vast majority of transport pathways between cell nucleus and cytoplasm. We identify here the translation elongation factor 1A (eEF1A) as the predominant nuclear export substrate of RanBP21/exportin 5 (Exp5). This cargo-exportin interaction is rather un usual in that eEF1A binds the exportin not directly, but instead via aminoacylated tRNAs. Exp5 thus represents the second directly RNA-binding exportin and mediates tRNA export in parallel with exportin-t. It was suggested recently that 10-15% of the cellular translation would occur in the nucleus. Our data rule out such a scenario and instead suggest that nuclear translation is actively suppressed by the nuclear export machinery. We found that the vast majority of translation initiation factors (eIF2, eIF2B, eIF3, eIF4A1, eIF5 and eIF5B), all three elongation factors (eEF1A, eEF1B and eEF2) and the termination factor eRF1 are strictly excluded from nuclei. Besides Exp5 and importin 13, CRM1 and as yet unidentified exportins also contribute to the depletion of translation factors from nuclei.
Figures






References
-
- Arts G.J., Fornerod,M. and Mattaj,I.W. (1998a) Identification of a nuclear export receptor for tRNA. Curr. Biol., 8, 305–314. - PubMed
-
- Battiste J.L., Pestova,T.V., Hellen,C.U. and Wagner,G. (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell, 5, 109–119. - PubMed
-
- Brogna S., Sato,T.A. and Rosbash,M. (2002) Ribosome components are associated with sites of transcription. Mol. Cell, 10, 93–104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

