Steroleosin, a sterol-binding dehydrogenase in seed oil bodies
- PMID: 11950969
- PMCID: PMC154248
- DOI: 10.1104/pp.010928
Steroleosin, a sterol-binding dehydrogenase in seed oil bodies
Erratum in
- Plant Physiol 2002 Aug;129(4):1930
Abstract
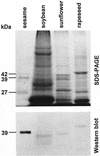
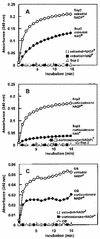
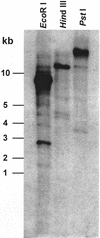
Besides abundant oleosin, three minor proteins, Sop 1, 2, and 3, are present in sesame (Sesamum indicum) oil bodies. The gene encoding Sop1, named caleosin for its calcium-binding capacity, has recently been cloned. In this study, Sop2 gene was obtained by immunoscreening, and it was subsequently confirmed by amino acid partial sequencing and immunological recognition of its overexpressed protein in Escherichia coli. Immunological cross recognition implies that Sop2 exists in seed oil bodies of diverse species. Along with oleosin and caleosin genes, Sop2 gene was transcribed in maturing seeds where oil bodies are actively assembled. Sequence analysis reveals that Sop2, tentatively named steroleosin, possesses a hydrophobic anchoring segment preceding a soluble domain homologous to sterol-binding dehydrogenases/reductases involved in signal transduction in diverse organisms. Three-dimensional structure of the soluble domain was predicted via homology modeling. The structure forms a seven-stranded parallel beta-sheet with the active site, S-(12X)-Y-(3X)-K, between an NADPH and a sterol-binding subdomain. Sterol-coupling dehydrogenase activity was demonstrated in the overexpressed soluble domain of steroleosin as well as in purified oil bodies. Southern hybridization suggests that one steroleosin gene and certain homologous genes may be present in the sesame genome. Comparably, eight hypothetical steroleosin-like proteins are present in the Arabidopsis genome with a conserved NADPH-binding subdomain, but a divergent sterol-binding subdomain. It is indicated that steroleosin-like proteins may represent a class of dehydrogenases/reductases that are involved in plant signal transduction regulated by various sterols.
Figures






References
-
- Altschul SF, Warren G, Webb M, Eugene WM, David JL. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Branden CI. Relation between structure and function of α/β proteins. Q Rev Biophys. 1980;13:317–338. - PubMed
-
- Breton R, Housset D, Mazza C, Fontecilla-Camps JC. The structure of a complex of human 17β-hydroxysteroid dehydrogenase with estradiol and NADP+identifies two principal targets for the design of inhibitors. Structure. 1996;15:905–915. - PubMed
-
- Chen ECF, Tai SSK, Peng CC, Tzen JTC. Identification of three novel unique proteins in seed oil bodies of sesame. Plant Cell Physiol. 1998;39:935–941. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

