Expression of full-length p53 and its isoform Deltap53 in breast carcinomas in relation to mutation status and clinical parameters
- PMID: 17054774
- PMCID: PMC1636663
- DOI: 10.1186/1476-4598-5-47
Expression of full-length p53 and its isoform Deltap53 in breast carcinomas in relation to mutation status and clinical parameters
Abstract
Background: The tumor suppressor gene p53 (TP53) controls numerous signaling pathways and is frequently mutated in human cancers. Novel p53 isoforms suggest alternative splicing as a regulatory feature of p53 activity.
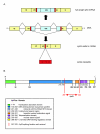
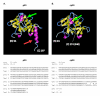
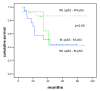
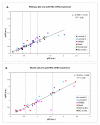
Results: In this study we have analyzed mRNA expression of both wild-type and mutated p53 and its respective Deltap53 isoform in 88 tumor samples from breast cancer in relation to clinical parameters and molecular subgroups. Three-dimensional structure differences for the novel internally deleted p53 isoform Deltap53 have been predicted. We confirmed the expression of Deltap53 mRNA in tumors using quantitative real-time PCR technique. The mRNA expression levels of the two isoforms were strongly correlated in both wild-type and p53-mutated tumors, with the level of the Deltap53 isoform being approximately 1/3 of that of the full-length p53 mRNA. Patients expressing mutated full-length p53 and non-mutated (wild-type) Deltap53, "mutational hybrids", showed a slightly higher frequency of patients with distant metastasis at time of diagnosis compared to other patients with p53 mutations, but otherwise did not differ significantly in any other clinical parameter. Interestingly, the p53 wild-type tumors showed a wide range of mRNA expression of both p53 isoforms. Tumors with mRNA expression levels in the upper or lower quartile were significantly associated with grade and molecular subtypes. In tumors with missense or in frame mutations the mRNA expression levels of both isoforms were significantly elevated, and in tumors with nonsense, frame shift or splice mutations the mRNA levels were significantly reduced compared to those expressing wild-type p53.
Conclusion: Expression of p53 is accompanied by the functionally different isoform Deltap53 at the mRNA level in cell lines and human breast tumors. Investigations of "mutational hybrid" patients highlighted that wild-type Deltap53 does not compensates for mutated p53, but rather may be associated with a worse prognosis. In tumors, both isoforms show strong correlations in different mutation-dependent mRNA expression patterns.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

