Clustered transcription factor genes regulate nicotine biosynthesis in tobacco
- PMID: 20959558
- PMCID: PMC2990138
- DOI: 10.1105/tpc.110.078543
Clustered transcription factor genes regulate nicotine biosynthesis in tobacco
Abstract
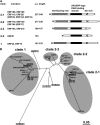
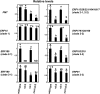
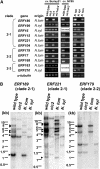
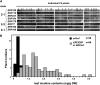
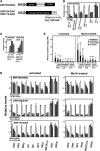
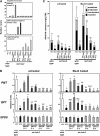
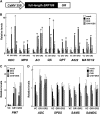
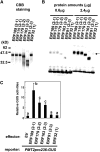
Tobacco (Nicotiana tabacum) synthesizes nicotine and related pyridine alkaloids in the root, and their synthesis increases upon herbivory on the leaf via a jasmonate-mediated signaling cascade. Regulatory NIC loci that positively regulate nicotine biosynthesis have been genetically identified, and their mutant alleles have been used to breed low-nicotine tobacco varieties. Here, we report that the NIC2 locus, originally called locus B, comprises clustered transcription factor genes of an ethylene response factor (ERF) subfamily; in the nic2 mutant, at least seven ERF genes are deleted altogether. Overexpression, suppression, and dominant repression experiments using transgenic tobacco roots showed both functional redundancy and divergence among the NIC2-locus ERF genes. These transcription factors recognized a GCC-box element in the promoter of a nicotine pathway gene and specifically activated all known structural genes in the pathway. The NIC2-locus ERF genes are expressed in the root and upregulated by jasmonate with kinetics that are distinct among the members. Thus, gene duplication events generated a cluster of highly homologous transcription factor genes with transcriptional and functional diversity. The NIC2-locus ERFs are close homologs of ORCA3, a jasmonate-responsive transcriptional activator of indole alkaloid biosynthesis in Catharanthus roseus, indicating that the NIC2/ORCA3 ERF subfamily was recruited independently to regulate jasmonate-inducible secondary metabolism in distinct plant lineages.
Figures


References
-
- Baldwin I.T. (1989). Mechanism of damage-induced alkaloid production in wild tobacco. J. Chem. Ecol. 15: 1661–1680 - PubMed
-
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: Chemical diversity in plant defense. Science 324: 746–748 - PubMed
-
- Cane K.A., Mayer M., Lidgett A.J., Michael A., Hamill J.D. (2005). Molecular analysis of alkaloid metabolism in AABB v. aabb genotype Nicotiana tabacum in response to wounding of aerial tissues and methyl jasmonate treatment of cultured roots. Funct. Plant Biol. 32: 305–320 - PubMed
-
- Chaplin J.F. (1975). Registration of LAFC 53 tobacco germplasm. Crop Sci. 15: 282
-
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

