The evolutionary value of recombination is constrained by genome modularity
- PMID: 16244707
- PMCID: PMC1262190
- DOI: 10.1371/journal.pgen.0010051
The evolutionary value of recombination is constrained by genome modularity
Abstract
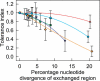
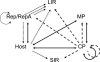
Genetic recombination is a fundamental evolutionary mechanism promoting biological adaptation. Using engineered recombinants of the small single-stranded DNA plant virus, Maize streak virus (MSV), we experimentally demonstrate that fragments of genetic material only function optimally if they reside within genomes similar to those in which they evolved. The degree of similarity necessary for optimal functionality is correlated with the complexity of intragenomic interaction networks within which genome fragments must function. There is a striking correlation between our experimental results and the types of MSV recombinants that are detectable in nature, indicating that obligatory maintenance of intragenome interaction networks strongly constrains the evolutionary value of recombination for this virus and probably for genomes in general.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Lehman NA. A case for the extreme antiquity of recombination. J Mol Evol. 2003;56:770–777. - PubMed
-
- Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. - PubMed
-
- Stemmer WPC. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

