Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA
- PMID: 11772617
- PMCID: PMC126540
- DOI: 10.1128/AEM.68.1.114-123.2002
Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA
Abstract
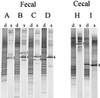
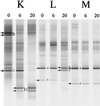
A Lactobacillus group-specific PCR primer, S-G-Lab-0677-a-A-17, was developed to selectively amplify 16S ribosomal DNA (rDNA) from lactobacilli and related lactic acid bacteria, including members of the genera Leuconostoc, Pediococcus, and WEISSELLA: Amplicons generated by PCR from a variety of gastrointestinal (GI) tract samples, including those originating from feces and cecum, resulted predominantly in Lactobacillus-like sequences, of which ca. 28% were most similar to the 16S rDNA of Lactobacillus ruminis. Moreover, four sequences of Leuconostoc species were retrieved that, so far, have only been detected in environments other than the GI tract, such as fermented food products. The validity of the primer was further demonstrated by using Lactobacillus-specific PCR and denaturing gradient gel electrophoresis (DGGE) of the 16S rDNA amplicons of fecal and cecal origin from different age groups. The stability of the GI-tract bacterial community in different age groups over various time periods was studied. The Lactobacillus community in three adults over a 2-year period showed variation in composition and stability depending on the individual, while successional change of the Lactobacillus community was observed during the first 5 months of an infant's life. Furthermore, the specific PCR and DGGE approach was tested to study the retention in fecal samples of a Lactobacillus strain administered during a clinical trial. In conclusion, the combination of specific PCR and DGGE analysis of 16S rDNA amplicons allows the diversity of important groups of bacteria that are present in low numbers in specific ecosystems to be characterized, such as the lactobacilli in the human GI tract.
Figures



References
-
- Ahrné, S., S. Nobaek, B. Jeppson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88–94. - PubMed
-
- Chagnaud, P., K. Machinis, L. A. Coutte, A. Marecat, and Y. Mercenier. 2001. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. J. Microbiol. Methods 44:139–148. - PubMed
-
- Collins, M. D., U. Rodrigues, M. Aguirre, J. A. E. Farrow, A. Martinez-Murcia, B. A. Philips, A. M. Williams, and S. Wallbanks. 1991. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett. 77:5–12.
Publication types
MeSH terms
Substances
Associated data
- GDB/AF335874
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

