Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean
- PMID: 11823184
- PMCID: PMC126663
- DOI: 10.1128/AEM.68.2.505-518.2002
Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean
Abstract
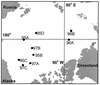
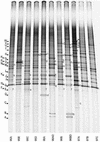
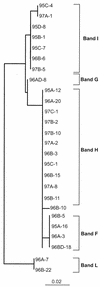
We analyzed the phylogenetic composition of bacterioplankton assemblages in 11 Arctic Ocean samples collected over three seasons (winter-spring 1995, summer 1996, and summer-fall 1997) by sequencing cloned fragments of 16S rRNA genes. The sequencing effort was directed by denaturing gradient gel electrophoresis (DGGE) screening of samples and the clone libraries. Sequences of 88 clones fell into seven major lineages of the domain Bacteria: alpha(36%)-, gamma(32%)-, delta(14%)-, and epsilon(1%)-Proteobacteria; Cytophaga-Flexibacter-Bacteroides spp. (9%); Verrucomicrobium spp. (6%); and green nonsulfur bacteria (2%). A total of 34% of the cloned sequences (excluding clones in the SAR11 and Roseobacter groups) had sequence similarities that were <94% compared to previously reported sequences, indicating the presence of novel sequences. DGGE fingerprints of the selected samples showed that most of the bands were common to all samples in all three seasons. However, additional bands representing sequences related to Cytophaga and Polaribacter species were found in samples collected during the summer and fall. Of the clones in a library generated from one sample collected in spring of 1995, 50% were the same and were most closely affiliated (99% similarity) with Alteromonas macleodii, while 50% of the clones in another sample were most closely affiliated (90 to 96% similarity) with Oceanospirillum sp. The majority of the cloned sequences were most closely related to uncultured, environmental sequences. Prominent among these were members of the SAR11 group. Differences between mixed-layer and halocline samples were apparent in DGGE fingerprints and clone libraries. Sequences related to alpha-Proteobacteria (dominated by SAR11) were abundant (52%) in samples from the mixed layer, while sequences related to gamma-proteobacteria were more abundant (44%) in halocline samples. Two bands corresponding to sequences related to SAR307 (common in deep water) and the high-G+C gram-positive bacteria were characteristic of the halocline samples.
Figures



References
-
- Aagaard, K., L. K. Coachman, and E. C. Carmack. 1981. On the halocline of the Arctic Ocean. Deep-Sea Res. 28:529-545.
-
- Aagaard, K., J. H. Swift, and E. C. Carmack. 1995. Thermohaline circulation in the Arctic Mediterranean Sea. J. Geophys. Res. 90:4833-4846.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

