RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse
- PMID: 11387227
- PMCID: PMC125495
- DOI: 10.1093/emboj/20.11.2943
RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse
Abstract
In mouse brain cDNA libraries generated from small RNA molecules we have identified a total of 201 different expressed RNA sequences potentially encoding novel small non-messenger RNA species (snmRNAs). Based on sequence and structural motifs, 113 of these RNAs can be assigned to the C/D box or H/ACA box subclass of small nucleolar RNAs (snoRNAs), known as guide RNAs for rRNA. While 30 RNAs represent mouse homologues of previously identified human C/D or H/ACA snoRNAs, 83 correspond to entirely novel snoRNAS: Among these, for the first time, we identified four C/D box snoRNAs and four H/ACA box snoRNAs predicted to direct modifications within U2, U4 or U6 small nuclear RNAs (snRNAs). Furthermore, 25 snoRNAs from either class lacked antisense elements for rRNAs or snRNAS: Therefore, additional snoRNA targets have to be considered. Surprisingly, six C/D box snoRNAs and one H/ACA box snoRNA were expressed exclusively in brain. Of the 88 RNAs not belonging to either snoRNA subclass, at least 26 are probably derived from truncated heterogeneous nuclear RNAs (hnRNAs) or mRNAS: Short interspersed repetitive elements (SINEs) are located on five RNA sequences and may represent rare examples of transcribed SINES: The remaining RNA species could not as yet be assigned either to any snmRNA class or to a part of a larger hnRNA/mRNA. It is likely that at least some of the latter will represent novel, unclassified snmRNAS:
Figures
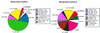

References
-
- Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 255–272.
-
- Bachellerie J.P., Cavaillé,J. and Qu,L.H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNAs revisited. In Garrett,R., Douthwaite,S., Liljas,A., Matheson,A., Moore,P.B. and Noller,H. (eds), Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203.
-
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. - PubMed
-
- Brosius J. (1999) Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica, 107, 209–238. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

