NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis
- PMID: 15316111
- PMCID: PMC520937
- DOI: 10.1105/tpc.104.023739
NAPP and PIRP encode subunits of a putative wave regulatory protein complex involved in plant cell morphogenesis
Erratum in
- Plant Cell. 2004 Nov;16(11):3168
Abstract
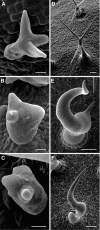
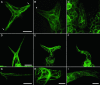
The ARP2/3 complex is an important regulator of actin nucleation and branching in eukaryotic organisms. All seven subunits of the ARP2/3 complex have been identified in Arabidopsis thaliana, and mutation of at least three of the subunits results in defects in epidermal cell expansion, including distorted trichomes. However, the mechanisms regulating the activity of the ARP2/3 complex in plants are largely unknown. In mammalian cells, WAVE and WASP proteins are involved in activation of the ARP2/3 complex. WAVE1 activity is regulated by a protein complex containing NAP1/HEM/KETTE/GEX-3 and PIR121/Sra-1/CYFIP/GEX-2. Here, we show that the WAVE1 regulatory protein complex is partly conserved in plants. We have identified Arabidopsis genes encoding homologs of NAP1 (NAPP), PIR121 (PIRP), and HSPC300 (BRK1). T-DNA inactivation of NAPP and PIRP results in distorted trichomes, similar to ARP2/3 complex mutants. The napp-1 mutant is allelic to the distorted mutant gnarled. The actin cytoskeleton in napp-1 and pirp-1 mutants shows orientation defects and increased bundling compared with wild-type plants. The results presented show that activity of the ARP2/3 complex in plants is regulated through an evolutionarily conserved mechanism.
Figures





References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Amann, K.J., and Pollard, T.D. (2001). The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3, 306–310. - PubMed
-
- Baumgartner, S., Martin, D., Chiquet-Ehrismann, R., Sutton, J., Desai, A., Huang, I., Kato, K., and Hromas, R. (1995). The HEM proteins: A novel family of tissue-specific transmembrane proteins expressed from invertebrates through mammals with an essential function in oogenesis. J. Mol. Biol. 251, 41–49. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad. Sci. III–Vie 316, 1194–1199.
-
- Blagg, S.L., Stewart, M., Sambles, C., and Insall, R.H. (2003). PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13, 1480–1487. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

