Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora
- PMID: 11679360
- PMCID: PMC93305
- DOI: 10.1128/AEM.67.11.5308-5314.2001
Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora
Abstract
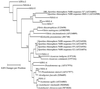
DNA was extracted from dry standing dead Spartina alterniflora stalks as well as dry Spartina wrack from the North Inlet (South Carolina) and Sapelo Island (Georgia) salt marshes. Partial nifH sequences were PCR amplified, the products were separated by denaturing gradient gel electrophoresis (DGGE), and the prominent DGGE bands were sequenced. Most sequences (109 of 121) clustered with those from alpha-Proteobacteria, and 4 were very similar (>99%) to that of Azospirillum brasilense. Seven sequences clustered with those from known gamma-Proteobacteria and five with those from known anaerobic diazotrophs. The diazotroph assemblages associated with dead Spartina biomass in these two salt marshes were very similar, and relatively few major lineages were represented.
Figures



References
-
- Affourtit J, Zehr J P, Paerl H W. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb Ecol. 2001;41:114–123. - PubMed
-
- Braun S T, Proctor L M, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–279.
-
- Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

