Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease
- PMID: 11854326
- PMCID: PMC150876
- DOI: 10.1172/JCI14099
Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease
Abstract
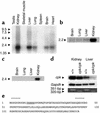
The congenital polycystic kidney (cpk) mutation is the most extensively characterized mouse model of polycystic kidney disease (PKD). The renal cystic disease is fully expressed in homozygotes and is strikingly similar to human autosomal recessive PKD (ARPKD), whereas genetic background modulates the penetrance of the corresponding defect in the developing biliary tree. We now describe the positional cloning, mutation analysis, and expression of a novel gene that is disrupted in cpk mice. The cpk gene is expressed primarily in the kidney and liver and encodes a hydrophilic, 145-amino acid protein, which we term cystin. When expressed exogenously in polarized renal epithelial cells, cystin is detected in cilia, and its expression overlaps with polaris, another PKD-related protein. We therefore propose that the single epithelial cilium is important in the functional differentiation of polarized epithelia and that ciliary dysfunction underlies the PKD phenotype in cpk mice.
Figures

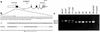

References
-
- Gabow P. Autosomal polycystic kidney disease. N Engl J Med. 1993;329:332–342. - PubMed
-
- Watnick T, Germino G. Molecular basis of autosomal dominant polycystic kidney disease. Semin Nephrol. 1999;19:327–343. - PubMed
-
- Guay-Woodford, L., Jafri, Z., and Bernstein, J. 1999. Other cystic diseases. In Comprehensive clinical nephrology. R. Johnson and J. Feehally, editors. Mosby International. London, United Kingdom. 50.1–50.2.
-
- Zerres K, et al. Mapping of the gene for autosomal recessive polycystic kidney disease (ARPKD) to chromosome 6p21-cen. Nat Genet. 1994;7:429–432. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

