Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity
- PMID: 11807059
- PMCID: PMC134827
- DOI: 10.1128/jb.184.4.992-1002.2002
Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity
Abstract
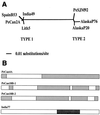
A new member of the IS605 transposable element family, designated ISHp608, was found by subtractive hybridization in Helicobacter pylori. Like the three other insertion sequences (ISs) known in this gastric pathogen, it contains two open reading frames (orfA and orfB), each related to putative transposase genes of simpler (one-gene) elements in other prokaryotes; orfB is also related to the Salmonella virulence gene gipA. PCR and hybridization tests showed that ISHp608 is nonrandomly distributed geographically: it was found in 21% of 194 European and African strains, 14% of 175 Bengali strains, 43% of 131 strains from native Peruvians and Alaska natives, but just 1% of 223 East Asian strains. ISHp608 also seemed more abundant in Peruvian gastric cancer strains than gastritis strains (9 of 14 versus 15 of 45, respectively; P = 0.04). Two ISHp608 types differing by approximately 11% in DNA sequence were identified: one was widely distributed geographically, and the other was found only in Peruvian and Alaskan strains. Isolates of a given type differed by < or = 2% in DNA sequence, but several recombinant elements were also found. ISHp608 marked with a resistance gene was found to (i) transpose in Escherichia coli; (ii) generate simple insertions during transposition, not cointegrates; (iii) insert downstream of the motif 5"-TTAC without duplicating target sequences; and (iv) require orfA but not orfB for its transposition. ISHp608 represents a widespread family of novel chimeric mobile DNA elements whose further analysis should provide new insights into transposition mechanisms and into microbial population genetic structure and genome evolution.
Figures




References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. - PubMed
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A., Smith, and K. A. Struhl. 1994. Current protocols in molecular biology, supplement 27, page 2.4.1. Greene Publishing and Wiley Interscience, New York, N.Y.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

