Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion
- PMID: 11731495
- PMCID: PMC311227
- DOI: 10.1101/gr.194701
Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion
Abstract
LINE-1 (L1) elements play an important creative role in genomic evolution by distributing both L1 and non-L1 DNA in a process called retrotransposition. A large percentage of the human genome consists of DNA that has been dispersed by the L1 transposition machinery. L1 elements are not randomly distributed in genomic DNA but are concentrated in regions with lower GC content. In an effort to understand the consequences of L1 insertions, we have begun an investigation of their genomic characteristics and the changes that occur to them over time. We compare human L1 insertions that were created either during recent human evolution or during the primate radiation. We report that L1 insertions are an important source for the creation of new microsatellites. We provide evidence that L1 first strand cDNA synthesis can occur from an internal priming event. We note that in contrast to older L1 insertions, recent L1s are distributed randomly in genomic DNA, and the shift in the L1 genomic distribution occurs relatively rapidly. Taken together, our data indicate that strong forces act on newly inserted L1 retrotransposons to alter their structure and distribution.
Figures
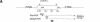
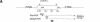

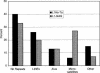

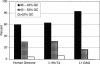

References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arcot SS, Adamson AW, Risch GW, LaFleur J, Robichaux MB, Lamerdin JE, Carrano AV, Batzer MA. High-resolution cartography of recently integrated human chromosome 19—specific Alu fossils. J Mol Biol. 1998;281:843–856. - PubMed
-
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
