Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure
- PMID: 12297986
- PMCID: PMC419997
- DOI: 10.1086/342928
Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure
Abstract
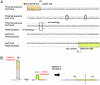
It is widely believed that at least three nonoverlapping regions of the human Y chromosome-AZFa, AZFb, and AZFc ("azoospermia factors" a, b, and c)-are essential for normal spermatogenesis. These intervals are defined by interstitial Y-chromosome deletions that impair or extinguish spermatogenesis. Deletion breakpoints, mechanisms, and lengths, as well as inventories of affected genes, have been elucidated for deletions of AZFa and of AZFc but not for deletions of AZFb or of AZFb plus AZFc. We studied three deletions of AZFb and eight deletions of AZFb plus AZFc, as assayed by the STSs defining these intervals. Guided by Y-chromosome sequence, we localized breakpoints precisely and were able to sequence nine of the deletion junctions. Homologous recombination can explain seven of these deletions but not the remaining two. This fact and our discovery of breakpoint hotspots suggest that factors in addition to homology underlie these deletions. The deletions previously thought to define AZFb were found to extend from palindrome P5 to the proximal arm of palindrome P1, 1.5 Mb within AZFc. Thus, they do not define a genomic region separate from AZFc. We also found that the deletions of AZFb plus AZFc, as assayed by standard STSs heretofore available, in fact extend from P5 to the distal arm of P1 and spare distal AZFc. Both classes of deletions are massive: P5/proximal-P1 deletions encompass up to 6.2 Mb and remove 32 genes and transcripts; P5/distal-P1 deletions encompass up to 7.7 Mb and remove 42 genes and transcripts. To our knowledge, these are the largest of all human interstitial deletions for which deletion junctions and complete intervening sequence are available. The restriction of the associated phenotype to spermatogenic failure indicates the remarkable functional specialization of the affected regions of the Y chromosome.
Figures





References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for previously published STSs sY105 [accession number G11994], sY116 [accession number G66528], sY117 [accession number G11996], sY118 [accession number G66529], sY127 [accession number G11998], sY143 [accession number G38347], sY149 [accession number G73322], sY254 [accession number G38349], sY579 [accession number G63909], sY639 [accession number G67162], sY1190 [accession number G67165], sY1192 [accession number G67166], sY1197 [accession number G67168], sY1201 [accession number G67170], and sY1206 [accession number G67171]; for new STSs, sY1207 [accession number G72341], sY1224 [accession number G72342], sY1227 [accession number G72343], sY1228 [accession number G72344], sY1257 [accession number G72345], sY1264 [accession number G72346], sY1289 [accession number G73323], sY1290 [accession number G73324], and sY1291 [accession number G72340]; for deletion junctions in AMC0110 [accession number AF395664], WHT3516 [accession number AF395665], WHT3642 [accession number AF395666], WHT3410 [accession number AF395667], WHT2825 [accession number AF395668], WHT2943 [accession number AF395669], WHT4396 [accession number AF437293], WHT4426 [accession number AF480412], and WHT4486 [accession number AF480413]; for reference sequence of the euchromatic, male-specific region of the human Y-chromosome long arm [accession numbers NT_011875 and NT_011903]; and for genes and transcripts affected by P5/P1 deletions, CDY2 [accession number AF080598], XKRY [accession number AF000997], HSFY [accession number AF332226], CYorf14 [accession number AF119903], CYorf15A [accession number AF332224], CYorf15B [accession number AF332225], SMCY [accession number U52191], EIF1AY [accession number AF000987], RPS4Y2 [accession number AF497481], RBMY1 [accession number X76060], PRY [accession number AF000988], BPY2 [accession number AF000980], DAZ [accession number U21663], CDY1 [accession number AF000981], CSPG4LY [accession number AF332228], GOLGA2LY [accession number AF332229], TTTY9 [accession number AF332238], TTTY14 [accession number AF332243], TTTY10 [accession number AF332239], TTTY13 [accession number AF332242], TTTY6 [accession number AF332237], TTTY5 [accession number AF332236], TTTY4 [accession number AF332231], and TTTY3 [accession number AF332230])
-
- The Human Y Chromosome: Annotated Sequence of the AZFc Palindromic Complex, http://staffa.wi.mit.edu/page/Y/azfc/ (for dot-plot code)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AZFc [MIM 400024] and Smith-Magenis syndrome [MIM 182290])
References
-
- Aradhya S, Bardaro T, Galgoczy P, Yamagata T, Esposito T, Patlan H, Ciccodicola A, Munnich A, Kenwrick S, Platzer M, D'Urso M, Nelson DL (2001) Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum Mol Genet 10:2557–2567 - PubMed
-
- Brandell RA, Mielnik A, Liotta D, Ye Z, Veeck LL, Palermo GD, Schlegel PN (1998) AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod 13:2812–2815 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

