Gene order evolution and paleopolyploidy in hemiascomycete yeasts
- PMID: 12093907
- PMCID: PMC123130
- DOI: 10.1073/pnas.142101099
Gene order evolution and paleopolyploidy in hemiascomycete yeasts
Abstract
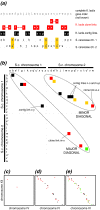
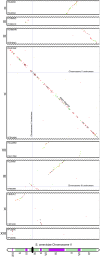
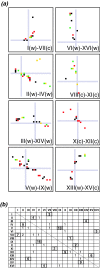
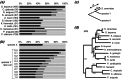
The wealth of comparative genomics data from yeast species allows the molecular evolution of these eukaryotes to be studied in great detail. We used "proximity plots" to visually compare chromosomal gene order information from 14 hemiascomycetes, including the recent Génolevures survey, to Saccharomyces cerevisiae. Contrary to the original reports, we find that the Génolevures data strongly support the hypothesis that S. cerevisiae is a degenerate polyploid. Using gene order information alone, 70% of the S. cerevisiae genome can be mapped into "sister" regions that tile together with almost no overlap. This map confirms and extends the map of sister regions that we constructed previously by using duplicated genes, an independent source of information. Combining gene order and gene duplication data assigns essentially the whole genome into sister regions, the largest gap being only 36 genes long. The 16 centromere regions of S. cerevisiae form eight pairs, indicating that an ancestor with eight chromosomes underwent complete doubling; alternatives such as segmental duplications can be ruled out. Gene arrangements in Kluyveromyces lactis and four other species agree quantitatively with what would be expected if they diverged from S. cerevisiae before its polyploidization. In contrast, Saccharomyces exiguus, Saccharomyces servazzii, and Candida glabrata show higher levels of gene adjacency conservation, and more cases of imperfect conservation, suggesting that they split from the S. cerevisiae lineage after polyploidization. This finding is confirmed by sequences around the C. glabrata TRP1 and IPP1 loci, which show that it contains sister regions derived from the same duplication event as that of S. cerevisiae.
Figures
Similar articles
-
Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi.Yeast. 1998 Mar 30;14(5):443-57. doi: 10.1002/(SICI)1097-0061(19980330)14:5<443::AID-YEA243>3.0.CO;2-L. Yeast. 1998. PMID: 9559552
-
Yeast genome duplication was followed by asynchronous differentiation of duplicated genes.Nature. 2003 Feb 20;421(6925):848-52. doi: 10.1038/nature01419. Nature. 2003. PMID: 12594514
-
Genomic exploration of the hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae.FEBS Lett. 2000 Dec 22;487(1):101-12. doi: 10.1016/s0014-5793(00)02289-4. FEBS Lett. 2000. PMID: 11152893
-
Evolutionary relationships between Saccharomyces cerevisiae and other fungal species as determined from genome comparisons.Rev Iberoam Micol. 2005 Dec;22(4):217-22. doi: 10.1016/s1130-1406(05)70046-2. Rev Iberoam Micol. 2005. PMID: 16499414 Review.
-
Evolutionary genomics: yeasts accelerate beyond BLAST.Curr Biol. 2004 May 25;14(10):R392-4. doi: 10.1016/j.cub.2004.05.015. Curr Biol. 2004. PMID: 15186766 Review.
Cited by
-
Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca.Genome Biol Evol. 2011;3:1150-63. doi: 10.1093/gbe/evr087. Epub 2011 Aug 22. Genome Biol Evol. 2011. PMID: 21859805 Free PMC article.
-
Three mating type-like loci in Candida glabrata.Eukaryot Cell. 2003 Apr;2(2):328-40. doi: 10.1128/EC.2.2.328-340.2003. Eukaryot Cell. 2003. PMID: 12684382 Free PMC article.
-
Local synteny and codon usage contribute to asymmetric sequence divergence of Saccharomyces cerevisiae gene duplicates.BMC Evol Biol. 2011 Sep 28;11:279. doi: 10.1186/1471-2148-11-279. BMC Evol Biol. 2011. PMID: 21955875 Free PMC article.
-
Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates.Genome Res. 2009 Aug;19(8):1404-18. doi: 10.1101/gr.086827.108. Epub 2009 May 13. Genome Res. 2009. PMID: 19439512 Free PMC article.
-
Modeling protein network evolution under genome duplication and domain shuffling.BMC Syst Biol. 2007 Nov 13;1:49. doi: 10.1186/1752-0509-1-49. BMC Syst Biol. 2007. PMID: 17999763 Free PMC article.
References
-
- Altmann-Jöhl R, Philippsen P. Mol Gen Genet. 1996;250:69–80. - PubMed
-
- Hartung K, Frishman D, Hinnen A, Wolfl S. Yeast. 1998;14:1327–1332. - PubMed
-
- Keogh R S, Seoighe C, Wolfe K H. Yeast. 1998;14:443–457. - PubMed
-
- Langkjaer R B, Nielsen M L, Daugaard P R, Liu W, Piskur J. J Mol Biol. 2000;304:271–288. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

