Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments
- PMID: 12663790
- PMCID: PMC152121
- DOI: 10.1128/jvi.77.8.4836-4847.2003
Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments
Abstract
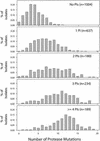
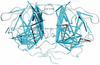
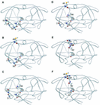
Although many human immunodeficiency virus type 1 (HIV-1)-infected persons are treated with multiple protease inhibitors in combination or in succession, mutation patterns of protease isolates from these persons have not been characterized. We collected and analyzed 2,244 subtype B HIV-1 isolates from 1,919 persons with different protease inhibitor experiences: 1,004 isolates from untreated persons, 637 isolates from persons who received one protease inhibitor, and 603 isolates from persons receiving two or more protease inhibitors. The median number of protease mutations per isolate increased from 4 in untreated persons to 12 in persons who had received four or more protease inhibitors. Mutations at 45 of the 99 amino acid positions in the protease-including 22 not previously associated with drug resistance-were significantly associated with protease inhibitor treatment. Mutations at 17 of the remaining 99 positions were polymorphic but not associated with drug treatment. Pairs and clusters of correlated (covarying) mutations were significantly more likely to occur in treated than in untreated persons: 115 versus 23 pairs and 30 versus 2 clusters, respectively. Of the 115 statistically significant pairs of covarying residues in the treated isolates, 59 were within 8 A of each other-many more than would be expected by chance. In summary, nearly one-half of HIV-1 protease positions are under selective drug pressure, including many residues not previously associated with drug resistance. Structural factors appear to be responsible for the high frequency of covariation among many of the protease residues. The presence of mutational clusters provides insight into the complex mutational patterns required for HIV-1 protease inhibitor resistance.
Figures

References
-
- Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300.
-
- Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. - PMC - PubMed
-
- Chen, Z., Y. Li, E. Chen, D. L. Hall, P. L. Darke, C. Culberson, J. A. Shafer, and L. C. Kuo. 1994. Crystal structure at 1.9-Å resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J. Biol. Chem. 269:26344-26348. - PubMed
-
- Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. - PMC - PubMed
-
- Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, and M. Shivaprakash. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

