Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis
- PMID: 11116097
- PMCID: PMC313064
- DOI: 10.1101/gr.10.12.2030
Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis
Abstract
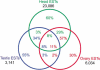
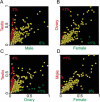
Identification and annotation of all the genes in the sequenced Drosophila genome is a work in progress. Wild-type testis function requires many genes and is thus of potentially high value for the identification of transcription units. We therefore undertook a survey of the repertoire of genes expressed in the Drosophila testis by computational and microarray analysis. We generated 3141 high-quality testis expressed sequence tags (ESTs). Testis ESTs computationally collapsed into 1560 cDNA set used for further analysis. Of those, 11% correspond to named genes, and 33% provide biological evidence for a predicted gene. A surprising 47% fail to align with existing ESTs and 16% with predicted genes in the current genome release. EST frequency and microarray expression profiles indicate that the testis mRNA population is highly complex and shows an extended range of transcript abundance. Furthermore, >80% of the genes expressed in the testis showed onefold overexpression relative to ovaries, or gonadectomized flies. Additionally, >3% showed more than threefold overexpression at p <0.05. Surprisingly, 22% of the genes most highly overexpressed in testis match Drosophila genomic sequence, but not predicted genes. These data strongly support the idea that sequencing additional cDNA libraries from defined tissues, such as testis, will be important tools for refined annotation of the Drosophila genome. Additionally, these data suggest that the number of genes in Drosophila will significantly exceed the conservative estimate of 13,601.
Figures
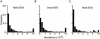
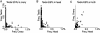



Comment in
-
Doubling the rewards: testis ESTs for Drosophila gene discovery and spermatogenesis expression profile analysis.Genome Res. 2000 Dec;10(12):1841-2. doi: 10.1101/gr.169400. Genome Res. 2000. PMID: 11116081 Review. No abstract available.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. - PubMed
-
- Ashburner M. A biologist's view of the Drosophila genome annotation assessment project. Genome Res. 2000;10:391–393. - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
