Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates
- PMID: 22830599
- PMCID: PMC4053731
- DOI: 10.1186/gb-2012-13-7-r64
Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates
Abstract
Background: While Staphylococcus epidermidis is commonly isolated from healthy human skin, it is also the most frequent cause of nosocomial infections on indwelling medical devices. Despite its importance, few genome sequences existed and the most frequent hospital-associated lineage, ST2, had not been fully sequenced.
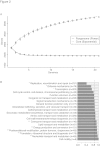
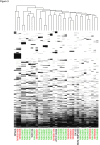
Results: We cultivated 71 commensal S. epidermidis isolates from 15 skin sites and compared them with 28 nosocomial isolates from venous catheters and blood cultures. We produced 21 commensal and 9 nosocomial draft genomes, and annotated and compared their gene content, phylogenetic relatedness and biochemical functions. The commensal strains had an open pan-genome with 80% core genes and 20% variable genes. The variable genome was characterized by an overabundance of transposable elements, transcription factors and transporters. Biochemical diversity, as assayed by antibiotic resistance and in vitro biofilm formation, demonstrated the varied phenotypic consequences of this genomic diversity. The nosocomial isolates exhibited both large-scale rearrangements and single-nucleotide variation. We showed that S. epidermidis genomes separate into two phylogenetic groups, one consisting only of commensals. The formate dehydrogenase gene, present only in commensals, is a discriminatory marker between the two groups.
Conclusions: Commensal skin S. epidermidis have an open pan-genome and show considerable diversity between isolates, even when derived from a single individual or body site. For ST2, the most common nosocomial lineage, we detect variation between three independent isolates sequenced. Finally, phylogenetic analyses revealed a previously unrecognized group of S. epidermidis strains characterized by reduced virulence and formate dehydrogenase, which we propose as a clinical molecular marker.
Figures





References
-
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;13:996–101. doi: 10.1086/591861. - DOI - PubMed
-
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;13:731–739. doi: 10.1016/S0140-6736(06)68231-7. - DOI - PubMed
Publication types
MeSH terms
Associated data
- BioProject/62343
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

