Positive selection driving diversification in plant secondary metabolism
- PMID: 16754868
- PMCID: PMC1482576
- DOI: 10.1073/pnas.0601738103
Positive selection driving diversification in plant secondary metabolism
Abstract
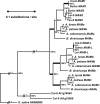
In Arabidopsis thaliana and related plants, glucosinolates are a major component in the blend of secondary metabolites and contribute to resistance against herbivorous insects. Methylthioalkylmalate synthases (MAM) encoded at the MAM gene cluster control an early step in the biosynthesis of glucosinolates and, therefore, are central to the diversification of glucosinolate metabolism. We sequenced bacterial artificial chromosomes containing the MAM cluster from several Arabidopsis relatives, conducted enzyme assays with heterologously expressed MAM genes, and analyzed MAM nucleotide variation patterns. Our results show that gene duplication, neofunctionalization, and positive selection provide the mechanism for biochemical adaptation in plant defense. These processes occur repeatedly in the history of the MAM gene family, indicating their fundamental importance for the evolution of plant metabolic diversity both within and among species.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

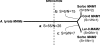
References
-
- Hartmann T., Kutchan T. M., Strack D. Phytochemistry. 2005;66:1193–1194. - PubMed
-
- Ehrlich P. R., Raven P. H. Evolution (Lawrence, Kans.) 1964;18:586–608.
-
- Windsor A. J., Reichelt M., Figuth A., Svatoš A., Kroymann J., Kliebenstein D. J., Gershenzon J., Mitchell-Olds T. Phytochemistry. 2005;66:1321–1333. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

