Evidence for existence of "mesotogas," members of the order Thermotogales adapted to low-temperature environments
- PMID: 16820506
- PMCID: PMC1489369
- DOI: 10.1128/AEM.00342-06
Evidence for existence of "mesotogas," members of the order Thermotogales adapted to low-temperature environments
Abstract
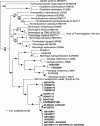
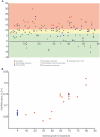
All cultivated isolates of the bacterial order Thermotogales are either thermophiles or hyperthermophiles, but Thermotogales 16S rRNA gene sequences have been detected in many mesophilic anaerobic and microaerophilic environments, particularly within communities involved in the remediation of pollutants. Here we provide metagenomic evidence for the existence of Thermotogales lineages, which we informally call "mesotoga," that are adapted to growth at lower temperatures. Two fosmid clones containing mesotoga DNA, originating from a low-temperature enrichment culture that degrades a polychlorinated biphenyl congener, were sequenced. Phylogenetic analysis clearly puts this bacterial lineage within the Thermotogales order, with the rRNA gene trees and 21 of 58 open reading frames strongly supporting this relationship. An analysis of protein sequence composition showed that mesotoga proteins are adapted to function at lower temperatures than are their identifiable homologs from thermophilic and hyperthermophilic members of the order Thermotogales, supporting the notion that this bacterium lives and grows optimally at lower temperatures. The phylogenetic analysis suggests that the mesotoga lineage from which our fosmids derive has used both the acquisition of genes from its neighbors and the modification of existing thermophilic sequences to adapt to a mesophilic lifestyle.
Figures

References
-
- Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. - PMC - PubMed
-
- Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

