Identification of sequence diversity in the Escherichia coli fliC genes encoding flagellar types H8 and H40 and its use in typing of Shiga toxin-producing E. coli O8, O22, O111, O174, and O179 strains
- PMID: 17135431
- PMCID: PMC1829044
- DOI: 10.1128/JCM.01627-06
Identification of sequence diversity in the Escherichia coli fliC genes encoding flagellar types H8 and H40 and its use in typing of Shiga toxin-producing E. coli O8, O22, O111, O174, and O179 strains
Abstract
Flagellar type H8 is associated with many strains of pathogenic Shiga toxin-producing Escherichia coli (STEC), such as O8, O22, O111, O174, and O179 strains. Serological typing of the H8 antigen is limited to motile strains only and suffers from cross-reactivity between flagellar H8 and H40 antigens. In order to develop a method useful for typing of motile and nonmotile STEC O111 and other strains, we have analyzed the flagellar antigen (fliC) genes in representative E. coli H8 and H40 types. Two genotypes of the fliC gene encoding H8 (the fliC-H8 gene) were identified. Genotype fliC-H8a was found to be conserved in STEC O111, O174, and O179 strains; and type fliC-H8b was associated with STEC O8 and O22 strains. Sequence variations were also found in the genetically closely related fliC-H40 gene, although the latter was not found to be associated with STEC strains. A PCR was developed for the specific identification of the fliC-H8 and the fliC-H40 genes in motile and nonmotile E. coli strains. Digestion of PCR products with HhaI resulted in restriction fragment length polymorphisms (RFLPs) which were associated with genotypes fliC-H8a and -H8b as well as with genotypes fliC-H40a and -H40b. The fliC-specific PCR/RFLP typing method was suitable for the rapid typing of motile and nonmotile STEC O8, O22, O111, O174, and O179 strains from different sources whose fliC-H8 genotypes were found to be highly conserved. The fliC genotyping method is advantageous over serotyping and is useful for epidemiological investigations and studies of the evolution of STEC clones.
Figures
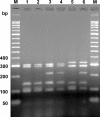
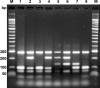
References
-
- Beutin, L., E. Strauch, S. Zimmermann, S. Kaulfuss, C. Schaudinn, A. Mannel, and H. R. Gelderblom. 2005. Genetical and functional investigation of fliC genes encoding flagellar serotype H4 in wildtype strains of Escherichia coli and in a laboratory E. coli K-12 strain expressing flagellar antigen type H48. BMC Microbiol. 5:4. - PMC - PubMed
-
- Blanco, J., M. Blanco, J. E. Blanco, A. Mora, M. P. Alonso, A. Gonzalez, and M. I. Bernadardez. 2001. Epidemiology of verocytotoxigenic Escherichia coli (VTEC) in ruminants, p. 113-148. In G. Duffy, P. Garvey, and D. A. McDowell (ed.), Verocytotoxigenic E. coli. Food and Nutrition Press, Inc., Trumbull, CT.
-
- Blanco, J. E., M. Blanco, M. P. Alonso, A. Mora, G. Dahbi, M. A. Coira, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from human patients: prevalence in Lugo, Spain, from 1992 through 1999. J. Clin. Microbiol. 42:311-319. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

