Sequence diversity, reproductive isolation and species concepts in Saccharomyces
- PMID: 16951060
- PMCID: PMC1602076
- DOI: 10.1534/genetics.106.062166
Sequence diversity, reproductive isolation and species concepts in Saccharomyces
Abstract
Using the biological species definition, yeasts of the genus Saccharomyces sensu stricto comprise six species and one natural hybrid. Previous work has shown that reproductive isolation between the species is due primarily to sequence divergence acted upon by the mismatch repair system and not due to major gene differences or chromosomal rearrangements. Sequence divergence through mismatch repair has also been shown to cause partial reproductive isolation among populations within a species. We have surveyed sequence variation in populations of Saccharomyces sensu stricto yeasts and measured meiotic sterility in hybrids. This allows us to determine the divergence necessary to produce the reproductive isolation seen among species. Rather than a sharp transition from fertility to sterility, which may have been expected, we find a smooth monotonic relationship between diversity and reproductive isolation, even as far as the well-accepted designations of S. paradoxus and S. cerevisiae as distinct species. Furthermore, we show that one species of Saccharomyces--S. cariocanus--differs from a population of S. paradoxus by four translocations, but not by sequence. There is molecular evidence of recent introgression from S. cerevisiae into the European population of S. paradoxus, supporting the idea that in nature the boundary between these species is fuzzy.
Figures

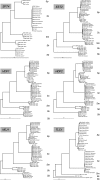

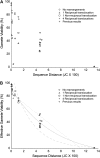


References
-
- Aa, E., J. P. Townsend, R. I. Adams, K. M. Nielsen and J. W. Taylor, 2006. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 702–715. - PubMed
-
- Cliften, P. F., L. W. Hillier, L. Fulton, T. Graves, T. Miner et al., 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11: 1175–1186. - PubMed
-
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

