Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples
- PMID: 17890331
- PMCID: PMC2168195
- DOI: 10.1128/AEM.01143-07
Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples
Abstract
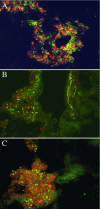
Little is known about bacterial communities that colonize mucosal surfaces in the human gastrointestinal tract, but they are believed to play an important role in host physiology. The objectives of this study were to investigate the compositions of these populations in the distal small bowel and colon. Healthy mucosal tissue from either the terminal ileum (n = 6) or ascending (n = 8), transverse (n = 8), or descending colon (n = 4) of 26 patients (age, 68.5 +/- 1.2 years [mean +/- standard deviation]) undergoing emergency resection of the large bowel was used to study these communities. Mucosa-associated eubacteria were characterized by using PCR-denaturing gradient gel electrophoresis (DGGE), while real-time PCR was employed for quantitative analysis. Mucosal communities were also visualized in situ using confocal laser scanning microscopy. DGGE banding profiles from all the gut regions exhibited at least 45% homology, with five descending colon profiles clustering at ca. 75% concordance. Real-time PCR showed that mucosal bacterial population densities were highest in the terminal ileum and that there were no significant differences in overall bacterial numbers in different parts of the colon. Bifidobacterial numbers were significantly higher in the large bowel than in the terminal ileum (P = 0.006), whereas lactobacilli were more prominent in the distal large intestine (P = 0.019). Eubacterium rectale (P = 0.0004) and Faecalibacterium prausnitzii (P = 0.001) were dominant in the ascending and descending colon. Site-specific colonization in the gastrointestinal tract may be contributory in the etiology of some diseases of the large intestine.
Figures


References
-
- Akkermans, A. D. L., E. G. Zoetendal, C. F. Favier, H. G. H. J. Heilig, W. M. Akkermans-van Vliet, and W. M. de Vos. 2000. Temperature and denaturing gradient gel electrophoresis analysis of 16S rRNA from human fecal samples. Biosci. Microflora 19:93-98.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anonymous. 2004. Cancer Statistics registrations; registrations of cancer diagnosed in 2001, England. Series MB1, no. 32. Office for National Statistics, London, United Kingdom.
-
- Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. T. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

