A rice Tc1/mariner-like element transposes in yeast
- PMID: 17041148
- PMCID: PMC1626630
- DOI: 10.1105/tpc.106.045906
A rice Tc1/mariner-like element transposes in yeast
Abstract
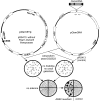
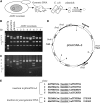
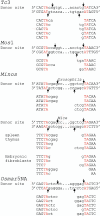
The Tc1/mariner transposable element superfamily is widely distributed in animal and plant genomes. However, no active plant element has been previously identified. Nearly identical copies of a rice (Oryza sativa) Tc1/mariner element called Osmar5 in the genome suggested potential activity. Previous studies revealed that Osmar5 encoded a protein that bound specifically to its own ends. In this report, we show that Osmar5 is an active transposable element by demonstrating that expression of its coding sequence in yeast promotes the excision of a nonautonomous Osmar5 element located in a reporter construct. Element excision produces transposon footprints, whereas element reinsertion occurs at TA dinucleotides that were either tightly linked or unlinked to the excision site. Several site-directed mutations in the transposase abolished activity, whereas mutations in the transposase binding site prevented transposition of the nonautonomous element from the reporter construct. This report of an active plant Tc1/mariner in yeast will provide a foundation for future comparative analyses of animal and plant elements in addition to making a new wide host range transposable element available for plant gene tagging.
Figures




References
-
- Auge-Gouillou, C., Hamelin, M.H., Demattei, M.V., Periquet, G., and Bigot, Y. (2001). The ITR binding domain of the Mariner Mos-1 transposase. Mol. Genet. Genomics 265 58–65. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

