Composition and structure of the centromeric region of rice chromosome 8
- PMID: 15037733
- PMCID: PMC412870
- DOI: 10.1105/tpc.019273
Composition and structure of the centromeric region of rice chromosome 8
Abstract
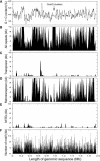
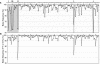
Understanding the organization of eukaryotic centromeres has both fundamental and applied importance because of their roles in chromosome segregation, karyotypic stability, and artificial chromosome-based cloning and expression vectors. Using clone-by-clone sequencing methodology, we obtained the complete genomic sequence of the centromeric region of rice (Oryza sativa) chromosome 8. Analysis of 1.97 Mb of contiguous nucleotide sequence revealed three large clusters of CentO satellite repeats (68.5 kb of 155-bp repeats) and >220 transposable element (TE)-related sequences; together, these account for approximately 60% of this centromeric region. The 155-bp repeats were tandemly arrayed head to tail within the clusters, which had different orientations and were interrupted by TE-related sequences. The individual 155-bp CentO satellite repeats showed frequent transitions and transversions at eight nucleotide positions. The 40 TE elements with highly conserved sequences were mostly gypsy-type retrotransposons. Furthermore, 48 genes, showing high BLAST homology to known proteins or to rice full-length cDNAs, were predicted within the region; some were close to the CentO clusters. We then performed a genome-wide survey of the sequences and organization of CentO and RIRE7 families. Our study provides the complete sequence of a centromeric region from either plants or animals and likely will provide insight into the evolutionary and functional analysis of plant centromeres.
Figures


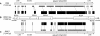
References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Bennetzen, J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42, 251–269. - PubMed
-
- Charlesworth, B., Sniegowski, P., and Stephan, W. (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

