Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka
- PMID: 16751340
- PMCID: PMC1484449
- DOI: 10.1101/gr.5016106
Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka
Abstract
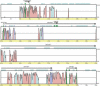
Sequencing of the human Y chromosome has uncovered the peculiarities of the genomic organization of a heterogametic sex chromosome of old evolutionary age, and has led to many insights into the evolutionary changes that occurred during its long history. We have studied the genomic organization of the medaka fish Y chromosome, which is one of the youngest heterogametic sex chromosomes on which molecular data are available. The Y specific and adjacent regions were sequenced and compared to the X. The male sex-determining gene, dmrt1bY, appears to be the only functional gene in the Y-specific region. The Y-specific region itself is derived from the duplication of a 43-kb fragment from linkage group 9. All other coduplicated genes except dmrt1bY degenerated. The Y-specific region has accumulated large stretches of repetitive sequences and duplicated pieces of DNA from elsewhere in the genome, thereby growing to 258 kb. Interestingly the non-recombining part of the Y did not spread out considerably from the original duplicated fragment, possibly because of a large sequence duplication bordering the Y-specific fragment. This may have conserved the more ancestral structure of the medaka Y and provides insights into some of the initial processes of Y chromosome evolution.
Figures
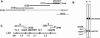







References
-
- Brunner B., Hornung U., Shan Z., Nanda I., Kondo M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Hornung U., Shan Z., Nanda I., Kondo M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Shan Z., Nanda I., Kondo M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Nanda I., Kondo M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Kondo M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Zend-Ajusch E., Haaf T., Ropers H.H., Shima A., Schmid M., Haaf T., Ropers H.H., Shima A., Schmid M., Ropers H.H., Shima A., Schmid M., Shima A., Schmid M., Schmid M., et al. Genomic organization and expression of the doublesex-related gene cluster in vertebrates and detection of putative regulatory regions for dmrt1. Genomics. 2001;77:8–17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
