Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation
- PMID: 15608331
- PMCID: PMC544494
- DOI: 10.1105/tpc.104.028381
Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation
Abstract
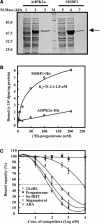
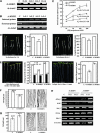
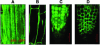
A putative Membrane Steroid Binding Protein (designated MSBP1) was identified and functionally characterized as a negative regulator of cell elongation in Arabidopsis thaliana. The MSBP1 gene encodes a 220-amino acid protein that can bind to progesterone, 5-dihydrotestosterone, 24-epi-brassinolide (24-eBL), and stigmasterol with different affinities in vitro. Transgenic plants overexpressing MSBP1 showed short hypocotyl phenotype and increased steroid binding capacity in membrane fractions, whereas antisense MSBP1 transgenic plants showed long hypocotyl phenotypes and reduced steroid binding capacity, indicating that MSBP1 negatively regulates hypocotyl elongation. The reduced cell elongation of MSBP1-overexpressing plants was correlated with altered expression of genes involved in cell elongation, such as expansins and extensins, indicating that enhanced MSBP1 affected a regulatory pathway for cell elongation. Suppression or overexpression of MSBP1 resulted in enhanced or reduced sensitivities, respectively, to exogenous progesterone and 24-eBL, suggesting a negative role of MSBP1 in steroid signaling. Expression of MSBP1 in hypocotyls is suppressed by darkness and activated by light, suggesting that MSBP1, as a negative regulator of cell elongation, plays a role in plant photomorphogenesis. This study demonstrates the functional roles of a steroid binding protein in growth regulation in higher plants.
Figures




References
-
- Alfandari, D., and Darribère, T. (1994). A simple PCR method for screening cDNA libraries. PCR Methods Appl. 4, 46–49. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Beato, M., Herrlich, P., and Schütz, G. (1995). Steroid hormone receptors: Many factors in search of a plot. Cell 83, 851–857. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

