In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs
- PMID: 10677516
- PMCID: PMC26493
- DOI: 10.1073/pnas.97.4.1665
In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs
Abstract
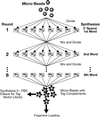
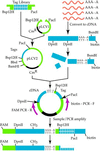
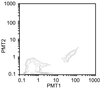
We describe a method for cloning nucleic acid molecules onto the surfaces of 5-micrometer microbeads rather than in biological hosts. A unique tag sequence is attached to each molecule, and the tagged library is amplified. Unique tagging of the molecules is achieved by sampling a small fraction (1%) of a very large repertoire of tag sequences. The resulting library is hybridized to microbeads that each carry approximately 10(6) strands complementary to one of the tags. About 10(5) copies of each molecule are collected on each microbead. Because such clones are segregated on microbeads, they can be operated on simultaneously and then assayed separately. To demonstrate the utility of this approach, we show how to label and extract microbeads bearing clones differentially expressed between two libraries by using a fluorescence-activated cell sorter (FACS). Because no prior information about the cloned molecules is required, this process is obviously useful where sequence databases are incomplete or nonexistent. More importantly, the process also permits the isolation of clones that are expressed only in given tissues or that are differentially expressed between normal and diseased states. Such clones then may be spotted on much more cost-effective, tissue- or disease-directed, low-density planar microarrays.
Figures
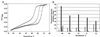

References
-
- Rowen L, Mahairas G, Hood L. Science. 1997;278:605–607. - PubMed
-
- Bishop J O, Morton J G, Rosbash M, Richardson M. Nature (London) 1974;250:199–204. - PubMed
-
- Ostrow R S, Woods W G, Vosika G J, Faras A J. Biochim Biophys Acta. 1979;562:92–102. - PubMed
-
- Lee C, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

