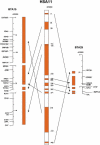
An ordered comparative map of the cattle and human genomes
- PMID: 10984454
- PMCID: PMC310912
- DOI: 10.1101/gr.145900
An ordered comparative map of the cattle and human genomes
Abstract
A cattle-human whole-genome comparative map was constructed using parallel radiation hybrid (RH) mapping in conjunction with EST sequencing, database mining for unmapped cattle genes, and a predictive bioinformatics approach (COMPASS) for targeting specific homologous regions. A total of 768 genes were placed on the RH map in addition to 319 microsatellites used as anchor markers. Of these, 638 had human orthologs with mapping data, thus permitting construction of an ordered comparative map. The large number of ordered loci revealed > or =105 conserved segments between the two genomes. The comparative map suggests that 41 translocation events, a minimum of 54 internal rearrangements, and repositioning of all but one centromere can account for the observed organizations of the cattle and human genomes. In addition, the COMPASS in silico mapping tool was shown to be 95% accurate in its ability to predict cattle chromosome location from random sequence data, demonstrating this tool to be valuable for efficient targeting of specific regions for detailed mapping. The comparative map generated will be a cornerstone for elucidating mammalian chromosome phylogeny and the identification of genes of agricultural importance."Ought we, for instance, to begin by discussing each separate species-in virtue of some common element of their nature, and proceed from this as a basis for the consideration of them separately?" from Aristotle, On the Parts of Animals, 350 B.C.E.
Figures
References
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Band M, Larson JH, Womack JE, Lewin HA. A radiation hybrid map of BTA23: Identification of a chromosomal rearrangement leading to separation of the cattle MHC class II subregions. Genomics. 1998;53:269–275. - PubMed
-
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, Williams JL, Sun HS, Eggen A, Agaba M, Aleyasin SA, et al. A medium-density genetic linkage map of the bovine genome. 1997. Mamm Genome. 1997;8:21–28. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials