Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site
- PMID: 11425735
- PMCID: PMC92994
- DOI: 10.1128/AEM.67.7.3149-3160.2001
Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site
Abstract
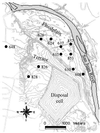
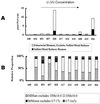
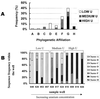
Microbially mediated reduction and immobilization of U(VI) to U(IV) plays a role in both natural attenuation and accelerated bioremediation of uranium-contaminated sites. To realize bioremediation potential and accurately predict natural attenuation, it is important to first understand the microbial diversity of such sites. In this paper, the distribution of sulfate-reducing bacteria (SRB) in contaminated groundwater associated with a uranium mill tailings disposal site at Shiprock, N.Mex., was investigated. Two culture-independent analyses were employed: sequencing of clone libraries of PCR-amplified dissimilatory sulfite reductase (DSR) gene fragments and phospholipid fatty acid (PLFA) biomarker analysis. A remarkable diversity among the DSR sequences was revealed, including sequences from delta-Proteobacteria, gram-positive organisms, and the Nitrospira division. PLFA analysis detected at least 52 different mid-chain-branched saturate PLFA and included a high proportion of 10me16:0. Desulfotomaculum and Desulfotomaculum-like sequences were the most dominant DSR genes detected. Those belonging to SRB within delta-Proteobacteria were mainly recovered from low-uranium (< or =302 ppb) samples. One Desulfotomaculum-like sequence cluster overwhelmingly dominated high-U (>1,500 ppb) sites. Logistic regression showed a significant influence of uranium concentration over the dominance of this cluster of sequences (P = 0.0001). This strong association indicates that Desulfotomaculum has remarkable tolerance and adaptation to high levels of uranium and suggests the organism's possible involvement in natural attenuation of uranium. The in situ activity level of Desulfotomaculum in uranium-contaminated environments and its comparison to the activities of other SRB and other functional groups should be an important area for future research.
Figures



References
-
- Abdelouas A, Lutze W, Gong W, Nuttall E H, Strietelmeier B A, Travis B J. Biological reduction of uranium in groundwater and subsurface soil. Sci Total Environ. 2000;250:21–35. - PubMed
-
- Abdelouas A, Leutz W, Nuttall E H. Uranium contamination in the subsurface: characterization and remediation. In: Burns P C, Finch R, editors. Uranium: mineralogy, geochemistry, and the environment. Reviews in mineralogy. Vol. 38. Washington, D.C.: Mineralogical Society of America; 1999. pp. 433–473.
-
- Agresti A. An introduction to categorical data analysis. New York, N.Y: John Wiley and Sons, Inc.; 1996.
-
- Brecklinghaus J, Schwartz W, Naveke R. Geomicrobiological studies. XIV. Heavy metal tolerance of desulfurizing bacteria under various ecological conditions. Z Allg Mikrobiol. 1981;21:65–76. - PubMed
-
- Canfield D E, DeMarias D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. - PubMed
Publication types
MeSH terms
Substances
Associated data
- GDB/AY015500
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

