Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities
- PMID: 11282596
- PMCID: PMC92760
- DOI: 10.1128/AEM.67.4.1494-1502.2001
Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities
Abstract
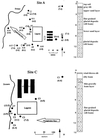
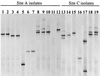
In this study, we used PCR typing methods to assess the presence of tetracycline resistance determinants conferring ribosomal protection in waste lagoons and in groundwater underlying two swine farms. All eight classes of genes encoding this mechanism of resistance [tet(O), tet(Q), tet(W), tet(M), tetB(P), tet(S), tet(T), and otrA] were found in total DNA extracted from water of two lagoons. These determinants were found to be seeping into the underlying groundwater and could be detected as far as 250 m downstream from the lagoons. The identities and origin of these genes in groundwater were confirmed by PCR-denaturing gradient gel electrophoresis and sequence analyses. Tetracycline-resistant bacterial isolates from groundwater harbored the tet(M) gene, which was not predominant in the environmental samples and was identical to tet(M) from the lagoons. The presence of this gene in some typical soil inhabitants suggests that the vector of antibiotic resistance gene dissemination is not limited to strains of gastrointestinal origin carrying the gene but can be mobilized into the indigenous soil microbiota. This study demonstrated that tet genes occur in the environment as a direct result of agriculture and suggested that groundwater may be a potential source of antibiotic resistance in the food chain.
Figures
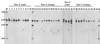

References
-
- Donoho A L. Biochemical studies on the fate of monensin in animals and in the environment. J Anim Sci. 1984;58:1528–1539. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

