Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior
- PMID: 11861899
- PMCID: PMC101233
- DOI: 10.1093/nar/30.5.1091
Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior
Abstract
Group II introns are novel genetic elements that have properties of both catalytic RNAs and retroelements. Initially identified in organellar genomes of plants and lower eukaryotes, group II introns are now being discovered in increasing numbers in bacterial genomes. Few of the newly sequenced bacterial introns are correctly identified or annotated by those who sequenced them. Here we have compiled and thoroughly analyzed group II introns and their fragments in bacterial DNA sequences reported to GenBank. Intron distribution in bacterial genomes differs markedly from the distribution in organellar genomes. Bacterial introns are not inserted into conserved genes, are often inserted outside of genes altogether and are frequently fragmented, suggesting a high rate of intron gain and loss. Some introns have multiple natural homing sites while others insert after transcriptional terminators. All bacterial group II introns identified to date encode reverse transcriptase open reading frames and are either active retroelements or derivatives of retroelements. Together, these observations suggest that group II introns in bacteria behave primarily as retroelements rather than as introns, and that the strategy for group II intron survival in bacteria is fundamentally different from intron survival in organelles.
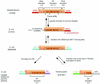
Figures


References
-
- van der Veen R., Arnbert,A.C., van der Horst,G., Bonen,L., Tabak,H.F. and Grivell,L.A. (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell, 44, 225–234. - PubMed
-
- Peebles C.L., Perlman,P.S., Mecklenburg,K.L., Petrillo,M.L., Tabor,J.H., Jarrell,K.A. and Cheng,H.-L. (1986) A self-splicing RNA excises an intron lariat. Cell, 44, 213–223. - PubMed
-
- Michel F. and Lang,B.F. (1985) Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature, 316, 641–643. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

