Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2
- PMID: 12676713
- PMCID: PMC154824
- DOI: 10.1128/AEM.69.4.2298-2305.2003
Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2
Abstract
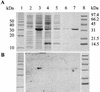
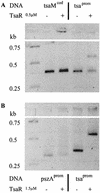
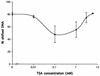
TsaR is the putative LysR-type regulator of the tsa operon (tsaMBCD) which encodes the first steps in the degradation of p-toluenesulfonate (TSA) in Comamonas testosteroni T-2. Transposon mutagenesis was used to knock out tsaR. The resulting mutant lacked the ability to grow with TSA and p-toluenecarboxylate (TCA). Reintroduction of tsaR in trans on an expression vector reconstituted growth with TSA and TCA. The tsaR gene was cloned into Escherichia coli with a C-terminal His tag and overexpressed as TsaR(His). TsaR(His) was subject to reversible inactivation by oxygen, which markedly influenced the experimental approaches used. Gel filtration showed TsaR(His) to be a monomer in solution. Overexpressed TsaR(His) bound specifically to three regions within the promoter between the divergently transcribed tsaR and tsaMBCD. The dissociation constant (K(D)) for the whole promoter region was about 0.9 micro M, and the interaction was a function of the concentration of the ligand TSA. A regulatory model for this LysR-type regulator is proposed on the basis of these data.
Figures


References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
-
- Behret, H., J. Ahlers, S. Ettel, E. Feicht, E. Futterer, I. Mangelsdorf, C. Pohlenz-Michel, H. Roβ, H. Sterzl-Eckert, D. Vogel, L. Weis, and K. Widmann. 1991. p-Toluolsulfonsäure, Beratergremium für umweltrelevante Alstoffe (BUA)-Stoffberichte, vol. 63. Verlag Chemie, Weinheim, Germany.
-
- Blatny, J. M., T. Brautaset, H. C. Winther Larsen, P. Karunakaran, and S. Valla. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35-51. - PubMed
-
- Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

